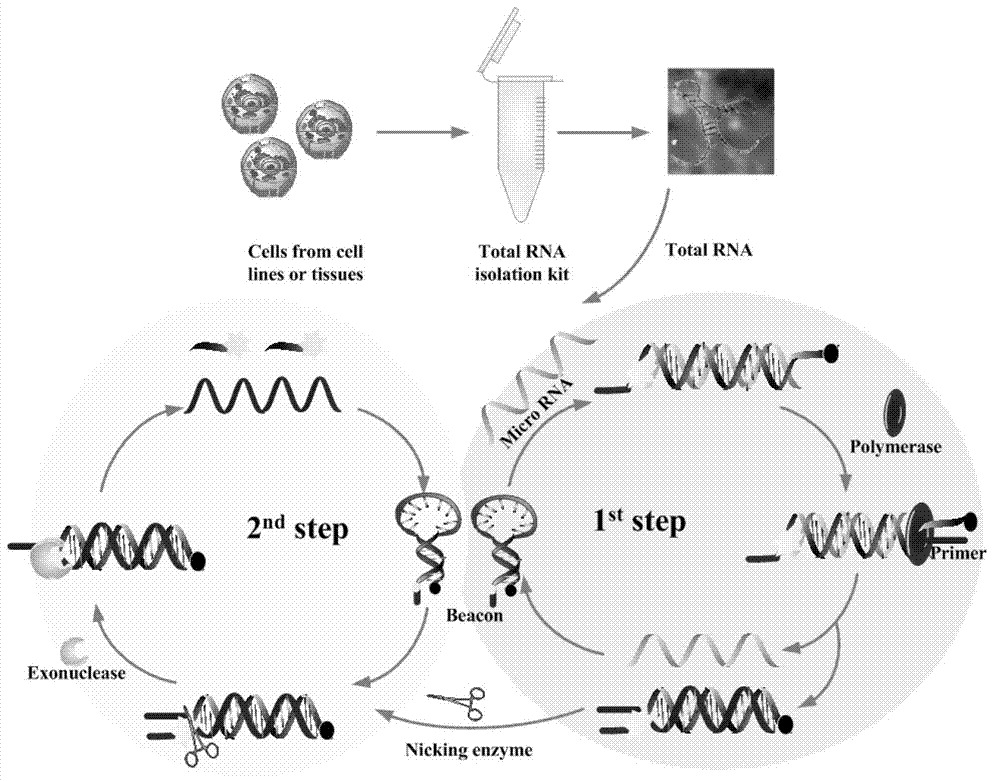
Testing method and kit for secondary circulation amplification of microRNA (Ribose Nucleic Acid)
A detection method, a quadratic technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of complex PCR design, short miRNAs sequence, and low sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1 uses miR-21 as a test object to further illustrate the present invention.
[0056] The enzymes and buffers used in this example include: NEB buffer 2, Bst large fragment DNA polymerase, lambda exonuclease, Nb.BbvCI nicking endonuclease, etc. were all purchased from NEB Company; molecular beacon probes, BSA, 8-base primers, miRNAs, DEPC water, dNTP, RNase inhibitor, etc. were purchased from Dalian Biotech.
[0057] The instrument used for fluorescence detection is a fluorescence spectrophotometer (Hitachi F-4500, Japan). Fluorescence spectrum measurement conditions: both the excitation and emission narrow peak widths are 5nm, the voltage PMT is 700v, the excitation wavelength is 490nm, and the emission wavelength scanning range is 500-600nm; a 250μL quartz cuvette is used for measurement.
[0058] The specific detection method is:
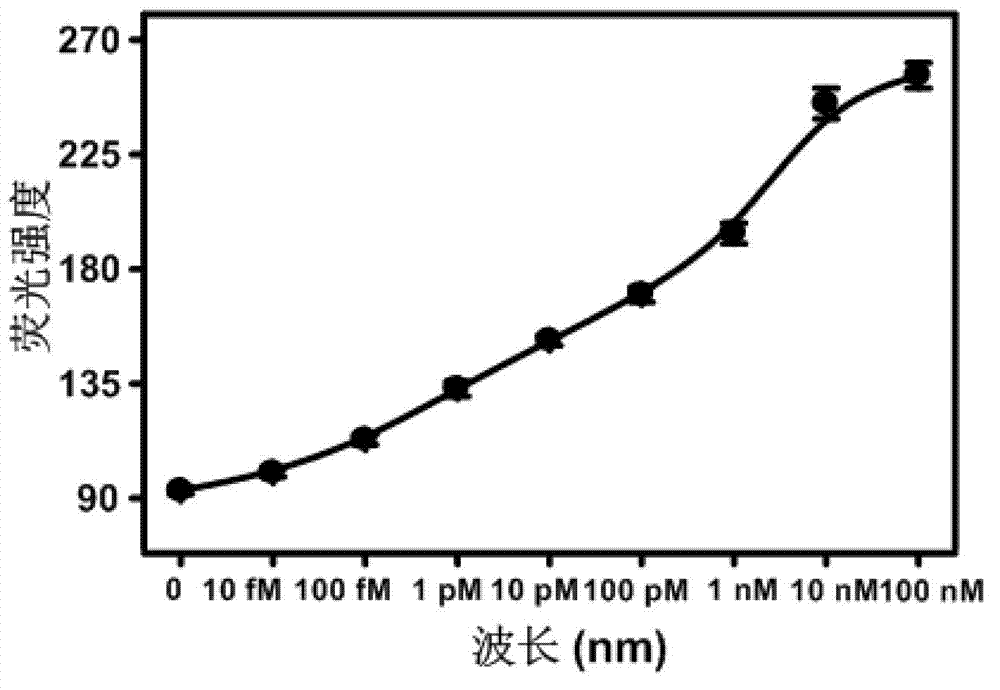
[0059] Step 1: Detection standard curve at 37°C: take 5-8 1.5ml centrifuge tubes, add molecular beacon probes with a final conce...
Embodiment 2
[0062] Example 2 uses the extract of breast cancer cells as the test substance to further illustrate the present invention.
[0063] Step 1: detection of breast cancer cells. Use mirVana first TM The kit extracts miRNA as the sample to be detected.
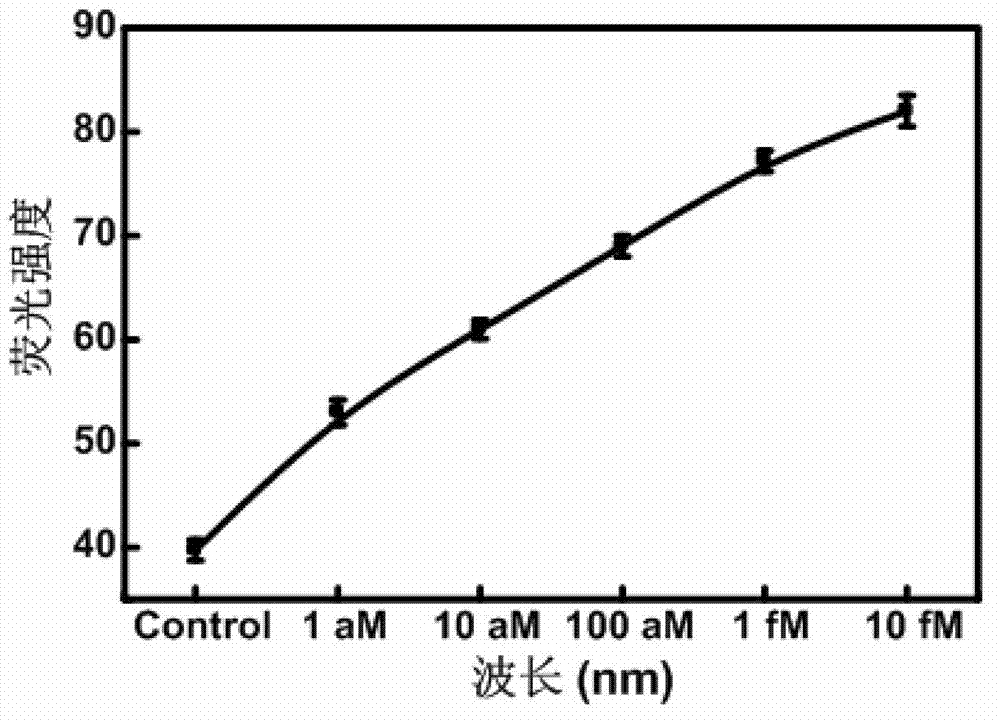
[0064] Step 2: Mix the sample to be tested with a final concentration of 500nM molecular beacon probe, 2μM 8-base primer, 400μM dNTPs, 16U Bst polymerase, 15U Nb.BbvCI nicking endonuclease, 12.5Ulambda exonuclease, Add BSA and RRI to a 1.5ml EP centrifuge tube. Detection was performed using a fluorometer. The breast cancer cell detection standard curve of cell number and fluorescence intensity can be obtained. (See Figure 4 ). The instrument setup parameters remain unchanged.
[0065] Step 3: We verified the extraction results of 1000 breast cancer cells. Using the above steps, we finally obtained a fluorescence value of 135. By comparing with the detection standard curve of breast cancer cells, it was found that 135 just...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com