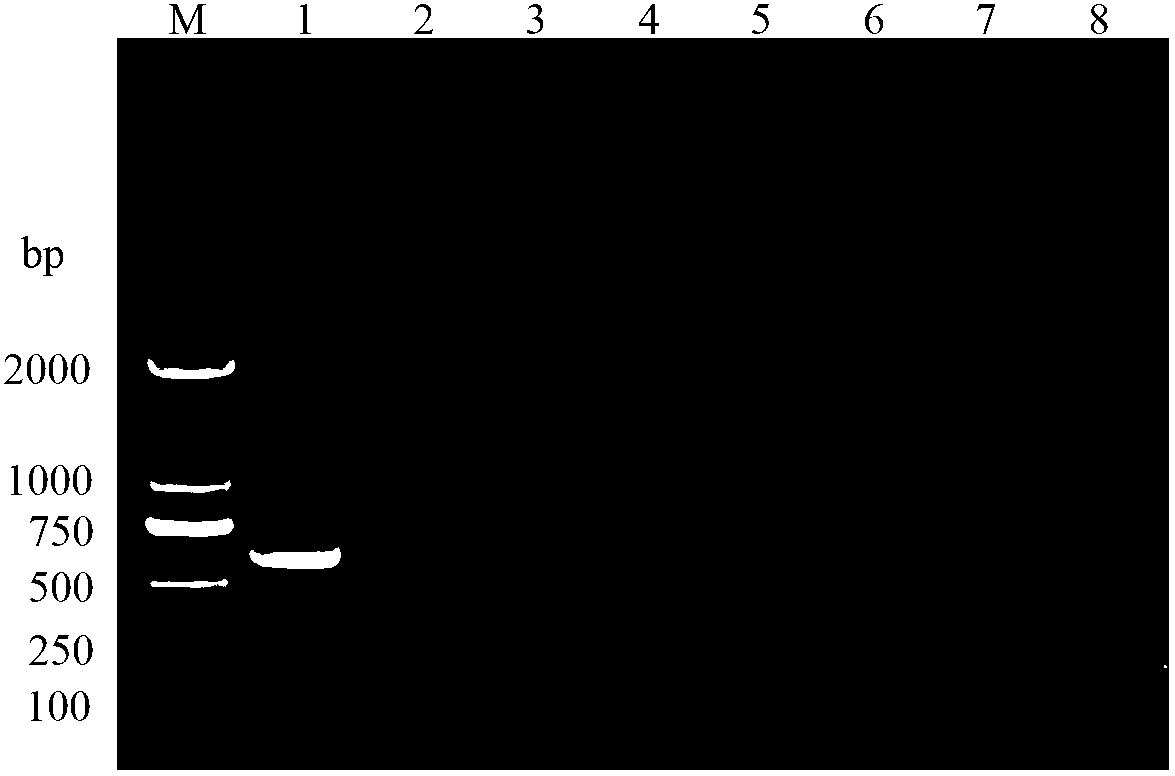Pair of phenacoccus solenopsis specific SS-COI primers, and rapid PCR detection method and kit
A technology for the detection method of fuso mealybug and its detection method, which is applied in the field of molecular biology, can solve the problems that there is no detection method for fuso mealybug, achieve the effect of meeting plant quarantine and pest monitoring/detection, simple operation process, and strong practicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Implementation Example 1: Amplification effect of primers PSZTF1 / PSZTR1 on Pyococcus hibiscus
[0028] 1) Preparation of mealybug template DNA
[0029] Place a single-headed mealybug (about 1 / 3 body length) on a parafilm membrane dripped with 20 μL of extraction buffer (50 mM Tris-HCl, lmMEDTA, 1% SDS, 20 mM NaCl, pH8.0), and use 0.2 mL of PCR Grind the bottom of the tube as a homogenizer, transfer the homogenate into a 1.5mL centrifuge tube with a micropipette; then wash the homogenizer and Prafilm membrane with 200 μL buffer solution for 4 times, transfer to the same centrifuge tube, mix well, and add 5 μL proteinase K (20mg / mL), mix thoroughly and place in a water bath at 60°C for 1.5h (mixing twice in the middle); then bathe in boiling water for 8min, add 220μL of chloroform / isoamyl alcohol (V:V=24:1) extract, gently After mixing dozens of times, place it on ice for 30 minutes; centrifuge at 4°C and 12000 r / min for 20 minutes, take about 200 μL of the supernatant a...
Embodiment 2
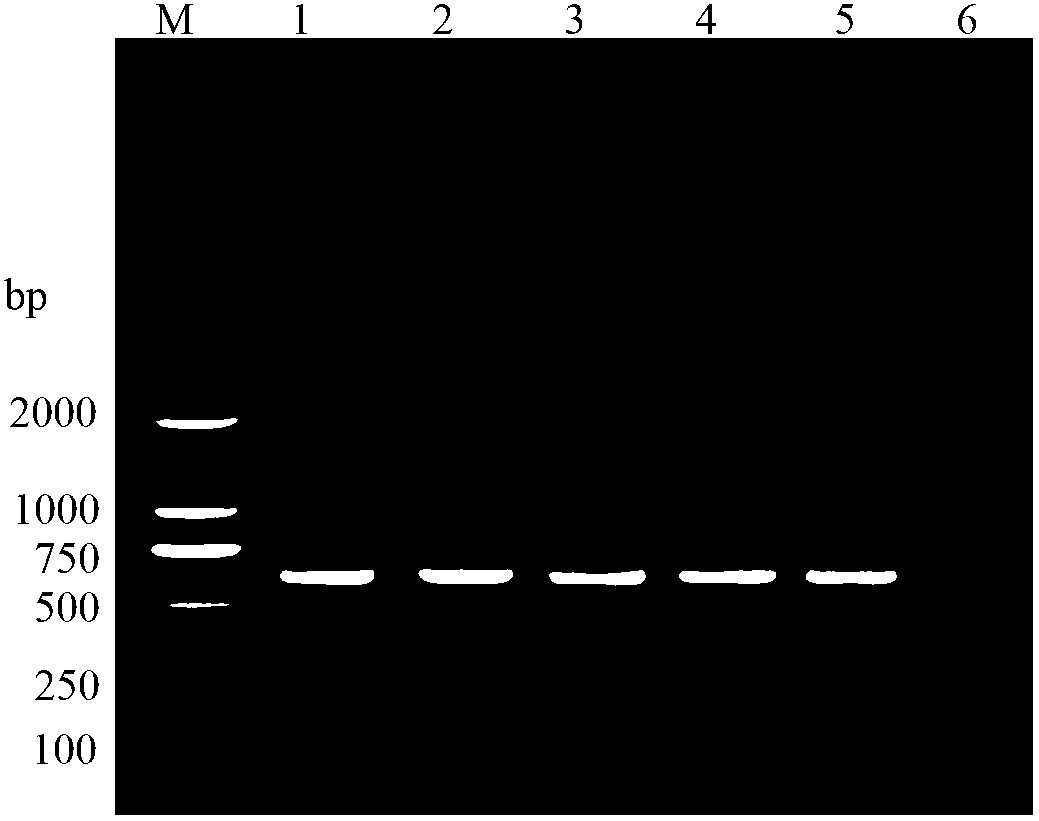
[0043] Implementation example 2: Amplification effect of primers PSZTF1 / PSZTR1 on different stages and ages of P. hibiscus
[0044] 1) Preparation of the template DNA of Pythococcus hibiscus cotton
[0045] Single-headed / single Pythococcus hibiscus of different stages and ages were placed on a parafilm dripped with 20 μL of extraction buffer (50 mM Tris-HCl, 1 mM EDTA, 1% SDS, 20 mM NaCl, pH 8.0). Use the bottom of a 0.2mL PCR tube as a homogenizer to grind thoroughly, transfer the homogenate into a 1.5mL centrifuge tube with a micropipette; then wash the homogenizer and Prafilm membrane with 200μL buffer solution 4 times, transfer to the same centrifuge tube, and mix well , add 5 μL of proteinase K (20mg / mL), mix thoroughly, and then place in a water bath at 60°C for 1.5 hours (mixing twice in the middle); then add 220 μL of chloroform / isoamyl alcohol (V:V=24:1) in a boiling water bath for 8 minutes After gently mixing the extract for dozens of times, place it on ice for 30 ...
Embodiment 3
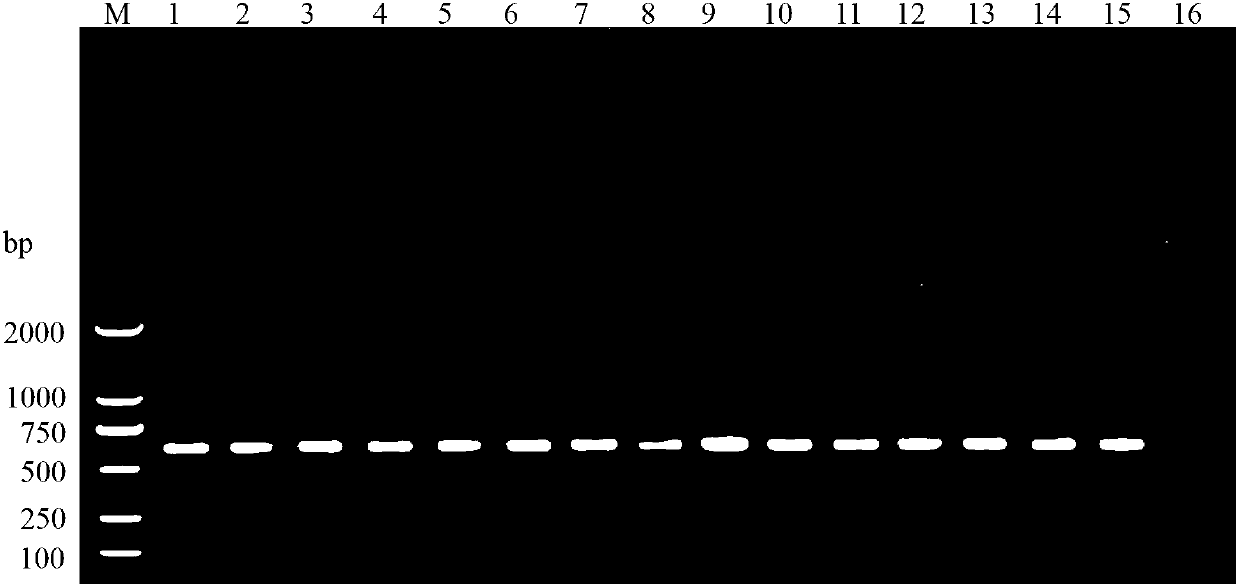
[0057] Implementation Example 3: Amplification results of primers PSZTF1 / PSZTR1 on Pythococcus hibiscus cotton collected from 14 different regions in 7 provinces and cities in my country and Pythias spongiosa from Pakistan intercepted by ports
[0058] 1) Preparation of the template DNA of Pythococcus hibiscus cotton
[0059] A single adult female P. solani (about 1 / 3 of the body length) was placed on a parafilm dripped with 20 μL of extraction buffer (50 mM Tris-HCl, 1 mM EDTA, 1% SDS, 20 mM NaCl, pH 8.0). The bottom of the 0.2mL PCR tube was used as a homogenizer to grind thoroughly, and the homogenate was transferred into a 1.5mL centrifuge tube with a micropipette; then the homogenizer and Prafilm membrane were washed with 200 μL buffer solution for 4 times, transferred to the same centrifuge tube, and mixed. Add 5 μL of proteinase K (20 mg / mL), mix thoroughly, and place in a 60°C water bath for 1.5 h (mixing twice in the middle); then add 220 μL of chloroform / isoamyl alco...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com