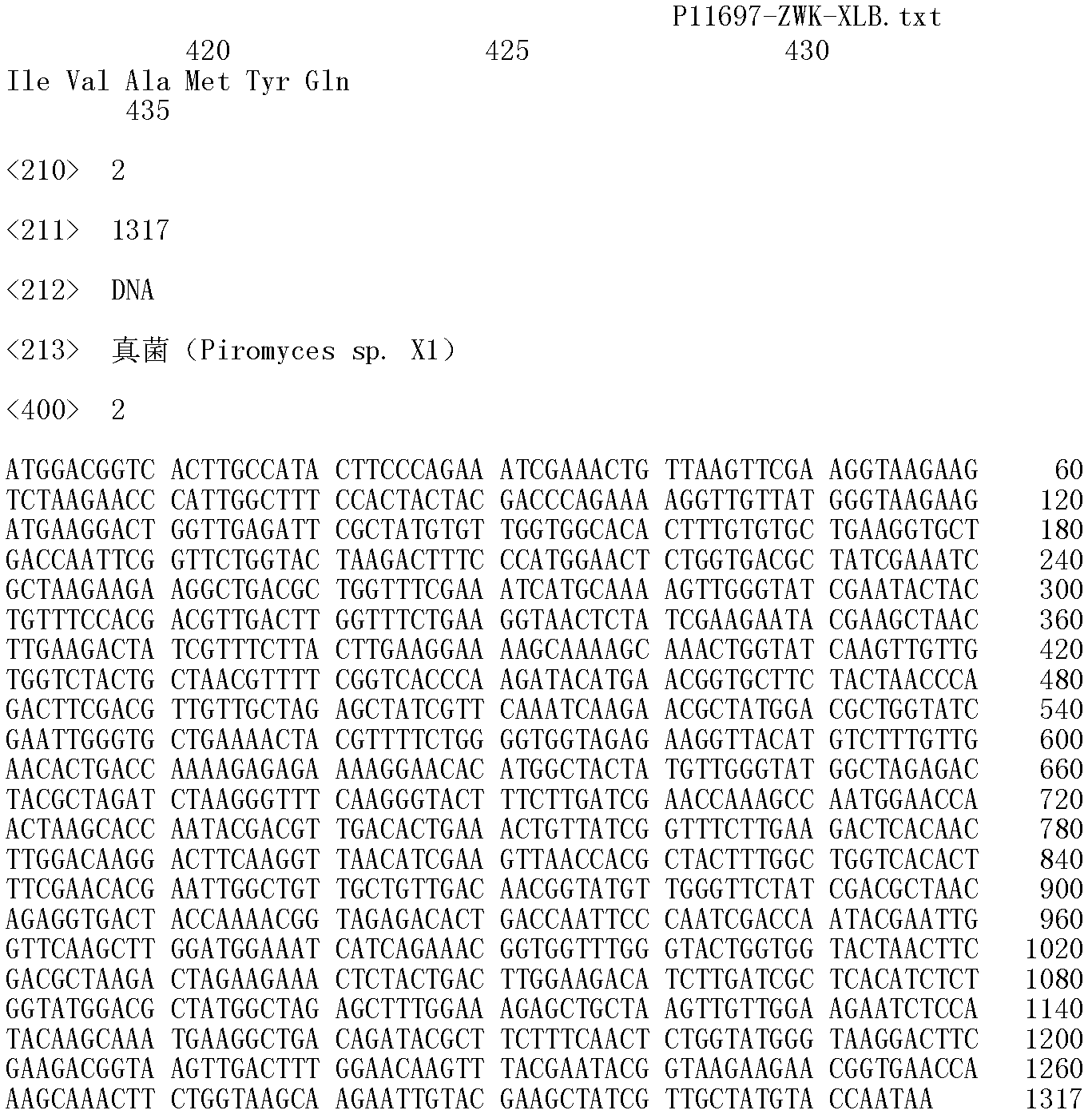
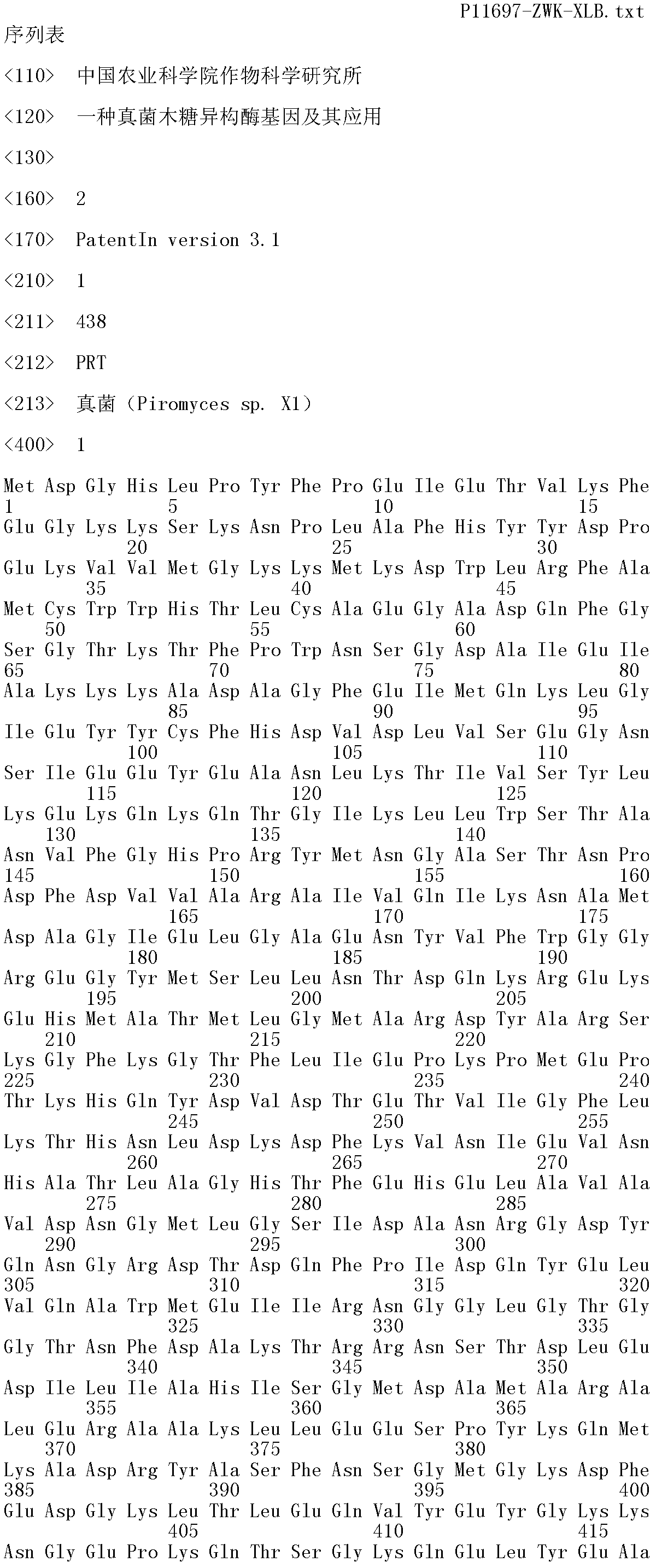Fungi xylose isomerase gene and application thereof
A technology of xylose isomerase and gene is applied in the application fields of recombinant plasmids and engineering strains, preparation of ethanol and other fermentation products, and can solve the problems of low activity and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Extraction of Piromyces sp.njau1 strain genome total DNA
[0038] ①Take about 0.5 g of Piromyces sp.njau1 strain and grind it into powder quickly in liquid nitrogen;
[0039] ②Add 4ml extract solution, shake and mix quickly;
[0040] ③Add an equal volume of 4ml of chloroform:isoamyl alcohol (24:1), and vortex for 3-5min;
[0041] ④10000rpm, 4°C, 5min;
[0042] ⑤The supernatant was extracted once more with volume of chloroform:isoamyl alcohol (24:1) (10000rpm, centrifuged at 4°C for 5min);
[0043] ⑥Take the supernatant, add 2 / 3 times the volume of -20°C pre-cooled isopropanol or 2.5 times the volume of absolute ethanol to precipitate, mix well, and let stand for about 30 minutes;
[0044] ⑦ Pick out the flocculent precipitate with a capillary glass rod, rinse repeatedly with 75% ethanol several times, then rinse once with absolute ethanol, blow dry, and resuspend in 500 μl TE;
[0045] ⑧Add 1μl RNaseA (10mg / ml), and treat at 37℃ for 1h;
[0046] ⑨ Extract...
Embodiment 2
[0048] Example 2 Cloning of Piromyces sp.njau1 xylose isomerase gene xylA
[0049] Using the total DNA of Piromyces sp.njau1 extracted in Example 1 as a template, PCR amplification was performed with primers designed with the conserved sequence of xylose isomerase. A fragment of about 900bp was obtained, which was recovered and connected with the pGEM-T easy vector and sent to Shanghai Sangon for sequencing.
[0050]According to the nucleotide sequence obtained by sequencing, three TAIL-PCR specific primers were designed upstream and downstream respectively: the design direction was the direction of the unknown region to be amplified, the position of sp2 was designed to be inside sp1, and sp3 was located inside sp2. The distance between every two primers is not strictly regulated, and the primer length is generally 22-30nt, and the annealing temperature is 60-65°C. And named them as usp1, usp2, usp3 (upstream specific primers), dsp1, dsp2, dsp3 (downstream specific primers) a...
Embodiment 3
[0054] Example 3 Construction of Xylose Isomerase Gene Expression Vector Plasmid
[0055] Primers were designed using the 5' and 3' end sequences of the gene encoding Piromyces sp.njau1 xylose isomerase, including EcoR I and Xba I sites. The PCR product was digested with EcoR I and Xba I. The final product was cloned into a vector generated by pYES2. In this vector, the GAL1 promoter on pYES2 was replaced with the TPI1 promoter to ensure constitutive expression of xylose isomerase, thereby eliminating the need for galactose in the medium. The TPI1 promoter was cloned from the Saccharomyces cerevisiae genome. This promoter was enzymatically cleaved into the Nhe I-EcoRI fragment. The TPI1 promoter and the PCR product of the gene encoding xylose isomerase were all connected to pYES2 cut with Spe I and Xba I to finally obtain the recombinant plasmid pYES-PXI containing the xylose isomerase gene.
[0056] Preparation of Escherichia coli Competent Cells and Transformation of Pla...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com