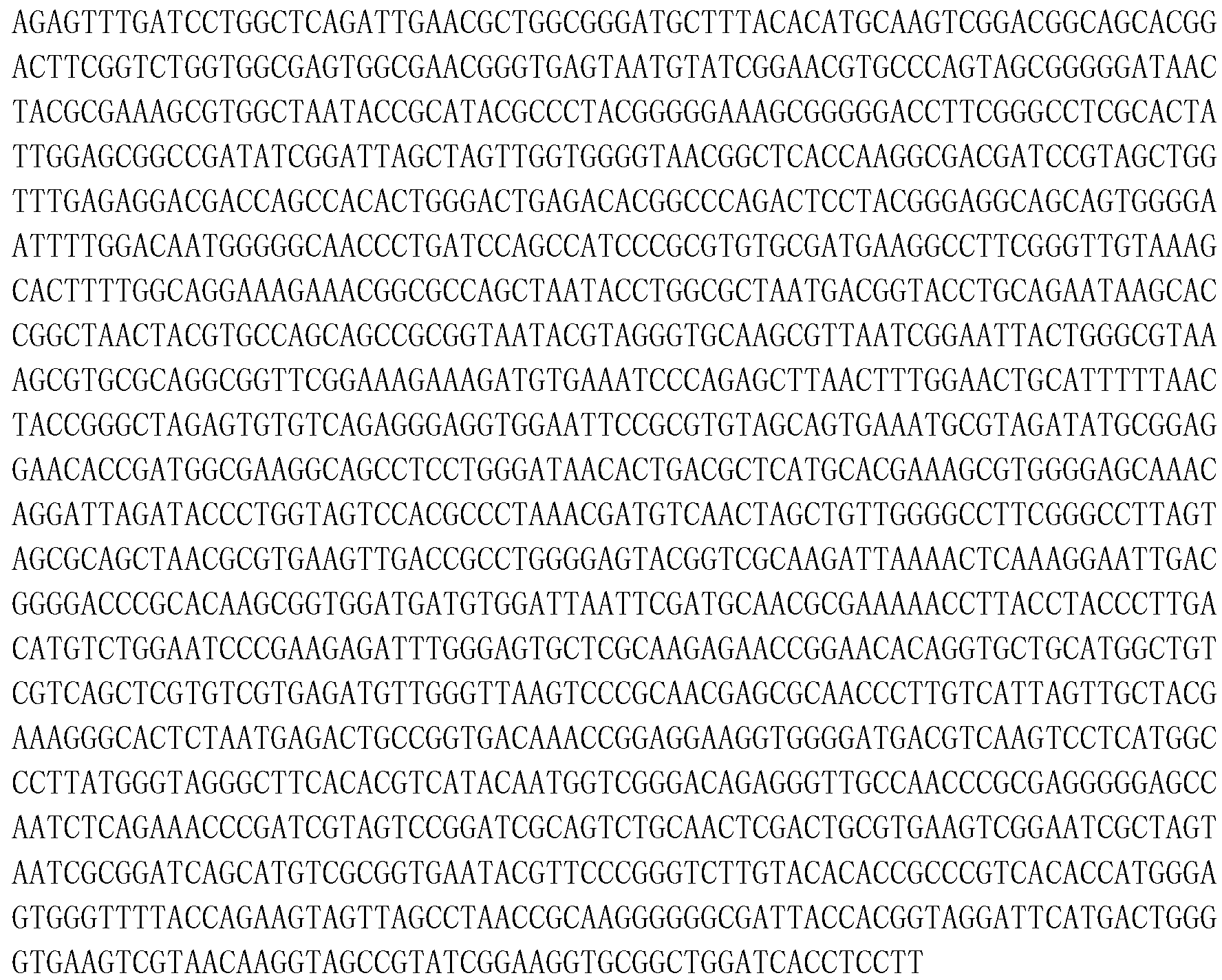6-aminopenicillanic acid degrading bacterium and screening method thereof
An aminopenicillanic acid, bacteria technology, applied in the direction of microorganism-based methods, bacteria, chemical instruments and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] The separation of the functional bacteria of embodiment 1 degradation 6-APA
[0025] Materials and Methods
[0026] 1. Medium
[0027] ① Enrichment medium:
[0028] Glucose, 20g; Peptone, 4g; Yeast powder, 1g; Beef extract, 2g; NaCl, 4g; K 2 HPO 4 , 1.5g; MgCl 2 ·6H 2 O, 0.1g; FeSO 4 ·7H 2 O, 0.1g; vitamin solution 10ml (cobalamin, 0.01g; ascorbic acid, 0.025g; riboflavin, 0.025g; citric acid, 0.02g; pyridoxal, 0.05g; folic acid, 0.01g; p-aminobenzoic acid , 0.01g; creatine, 0.025g); trace element solution (MnSO 4 ·7H 2 O, 0.01g; ZnSO 4 ·7H 2 O, 0.05g; H 3 BO 3 , 0.01g; N(CH 2 COOH) 3 , 4.5g; CaCl 2 2H 2 O, 0.01g; Na 2 MoO 4 , 0.01g; CoCl 2 ·6H2 O, 0.2g; AlK(SO 4 ) 2 , 0.01g) 10ml, finally dissolved in 1L.
[0029] ②Separation medium:
[0030] Glucose 10g; 6-APA 5g; NaCl, 4g; K 2 HPO 4 , 1.5g; MgCl 2 ·6H 2 O, 0.1g; FeSO 4 ·7H 2 O, 0.1g; vitamin solution 10ml (cobalamin, 0.01g; ascorbic acid, 0.025g; riboflavin, 0.025g; citric acid, 0.02g; ...
Embodiment 2
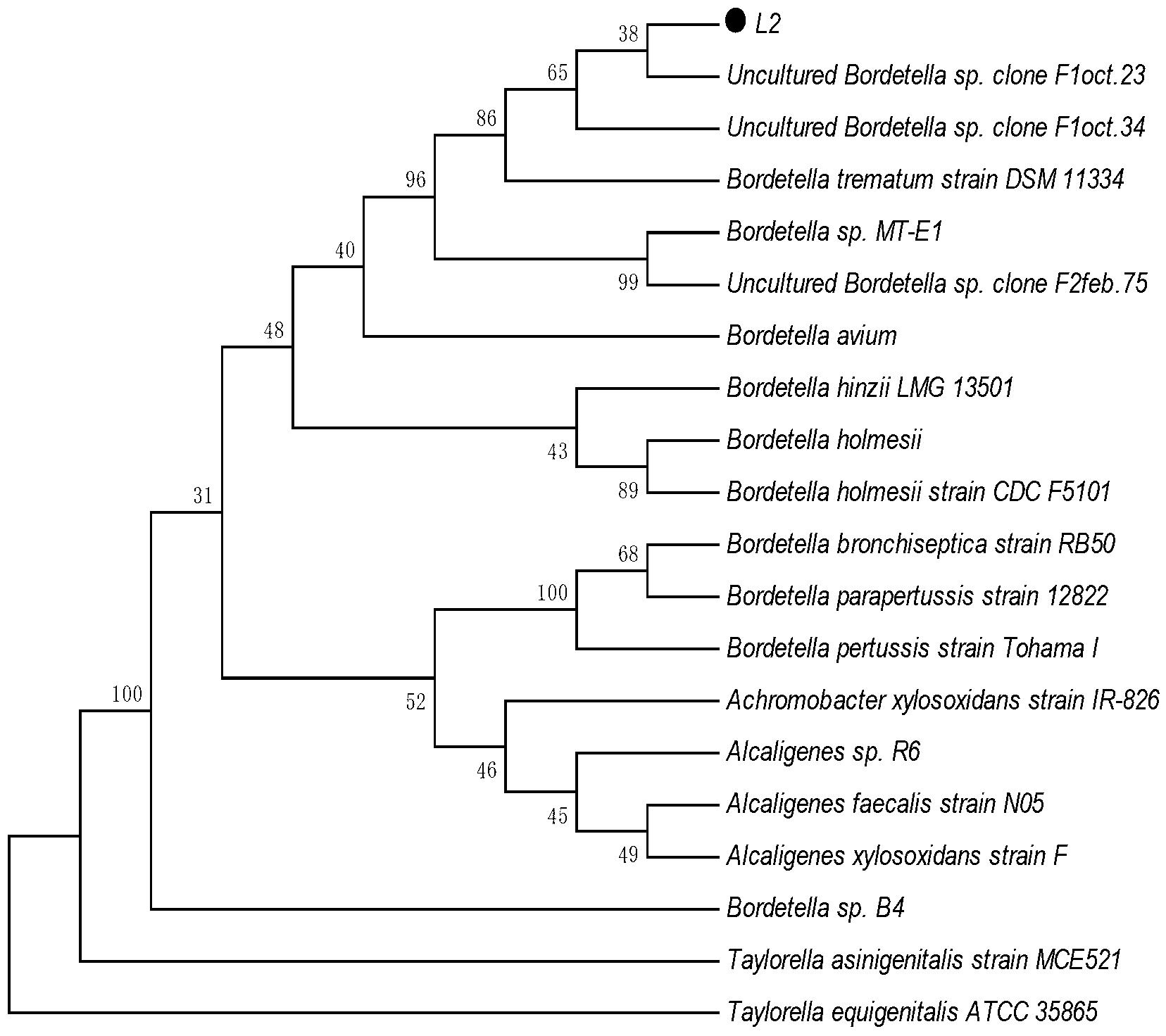
[0033] The strain identification of embodiment 2 bacterial strain L2
[0034] 1. Genomic DNA extraction
[0035] The above-mentioned strain L2 was mass-cultured according to conventional technical means, and then its genomic DNA was obtained.
[0036] 2. Identification method of 16S rDNA of strain L2
[0037] 2. PCR amplification and sequencing of 116S rRNA gene sequence and construction of phylogenetic tree
[0038] 2.1. PCR amplification of 116S rRNA gene sequence
[0039] The primers at both ends of the amplified 16S rRNA gene sequence are universal primers: forward primer BSF8 / 20: 5'-AGAGTTTGATCCTGGCTCAG-3' (SEQ ID NO: 1) and reverse primer BSR1541 / 20: 5'-AAGGAGGTGATCCAGCCGCA-3' (SEQ ID NO: 2). The PCR reaction system was 50 μL, and the reaction conditions were denaturation at 94°C for 5 minutes; followed by 35 cycle reactions: denaturation at 94°C for 45 s, annealing at 50°C for 45 s, and extension at 72°C for 90 s; then extension at 72°C for 10 min, and finally store...
Embodiment 36
[0073] The degradation rate of embodiment 36-APA
[0074] In this study, high performance liquid chromatography was used to determine the concentration of 6-APA. Agilent1100 high performance liquid chromatography of Agilent Technologies Co., Ltd. was used for determination. Chromatographic column: XDB-C18 (4.6×150mm, 5μm). The detection conditions are flow: 0.5mL / min, injection volume: 10μL, temperature: 30°C, wavelength: 210nm, 220nm, 230nm, 240nm, 260nm, mobile phase: methanol: water = 50:50.
[0075] Inoculate 0.1ml of strain L2 bacterial solution with 100ml of separation medium (the content of 6-APA is 300mg / L), culture at 37°C and 170rpm for 72hr, measure the concentration of 6-APA, and calculate its removal rate. The calculated removal rate of 6-APA in strain L2 was 28%.
[0076]
[0077]
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com