Method for fast and simple enrichment of trace quantity of animal tissue mitochondrial DNAs
An animal tissue and mitochondrial technology, applied in recombinant DNA technology, DNA preparation, etc., can solve the problems of large demand for samples, high cost, cumbersome process, etc., and achieves less initial tissue consumption, short experiment time, and equipment requirements. low effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Analysis of animal barcode label cox1 and mitochondrial 16S rRNA gene fragments in Macrobrachium rosenbergii muscle tissue
[0027] 1. Take a small piece of Macrobrachium rosenbergii tail muscle, put it in liquid nitrogen and grind it to powder, take about 20mg tissue sample powder, add 100μL extraction buffer, and mix with a vortex oscillator for 1min;
[0028] 2. Add 200 μL of alkaline lysate to the mixture, mix by inversion for 30 sec, and ice bath for 5 min;
[0029] 3. Add 150 μL 3M potassium acetate solution to the mixture, mix by inversion for 30 sec, and ice bath for 10 min;
[0030] 4. Add 0.45 μL of 25 mg / mL RNase A (Sangon Bioengineering Co., Ltd., product number R0675) to the mixture, incubate at 37°C for 1 hr, cool to room temperature naturally, centrifuge at 12,000×g for 10 min, and transfer the supernatant to a new 1.5 mL centrifuge tube;
[0031] 5. Add an equal volume of phenol (pH8.0) to the supernatant, mix at room temperature for 5 m...
Embodiment 2
[0042] Example 2: Analysis of individual barcode tags cox1 and mitochondrial 16S rRNA gene fragments of Artemia sinensis
[0043] 1. Take 4 Artemia sinensis adults (2 females and 2 males), put them in a 1.5mL centrifuge tube, weigh (20-30mg), add 100μL of extraction buffer, and grind on ice with a micro-grinding rod until uniform , vortex mixer for 1 min;
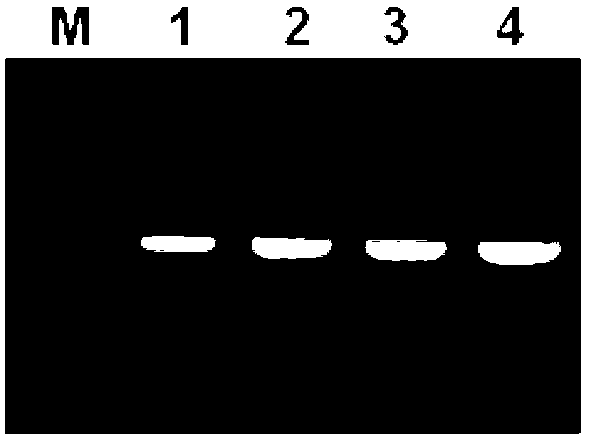
[0044] 2. Each tube was subjected to mitochondrial DNA enrichment, fragment amplification, and sequence analysis according to steps 2 to 12 of Example 1. For the results, see Figure 2 ~ Figure 3 and SEQ ID No.3-4.
[0045] SEQ ID No.3 is the cox1 sequence of Artemia sinensis (length 709bp, primer positions are underlined):
[0046] GGTCAACAAATCATAAAGATATTGG GACTTTATACTTTATTTTTGGAG CTTGAGCAGGAATAGTAGGAACTTCTTTAAGTATACTTATTCGAGCG GAATTAGGTCAACCTGGCTCCCTAATTGGAGATGAACAGGTATATAA TGTTATTGTAACAGCTCATGCATTTATTATAATTTTTTTCATAGTTATG CCTATCCTCATTGGAGGGTTTGGTAACTGATTAGTACCTATTATACTA GGAGCTCCTGATATGGCTTTTCCCCGCTTAAACAATTTAAGATTT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com