Application of miR-34a gene in non-small cell lung cancer
A non-small cell lung cancer and gene technology, applied in the direction of DNA / RNA fragments, recombinant DNA technology, drug combination, etc., can solve the problems of lack of early diagnosis and treatment methods for lung cancer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
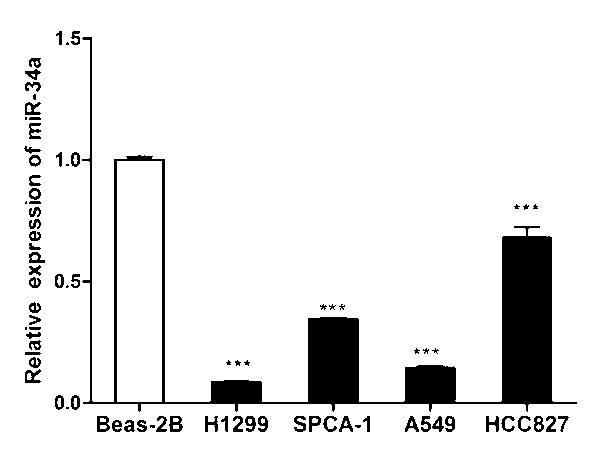
[0038] Example 1: According to the results of Solexa sequencing, it is determined that miR-34a is under-expressed in non-small cell lung cancer cell lines
[0039] The k-ras gene mutation mouse lung cancer model (L703T2) and the normal lung cancer model (L1805) used in the present invention are provided by the research group of Professor Ji Hongbin from the Institute of Biochemical Cells, Chinese Academy of Sciences.
[0040] The lung tissues of normal mice and those of mice with non-small cell lung cancer were sampled. The tissues were lysed by the TRIZOL method, and chloroform was added. After the protein and nucleic acid were layered, the supernatant was removed by centrifugation and isopropanol precipitation was added. After centrifugation again, the pellet was washed with ethanol and dried to obtain total RNA. TaKaRa company’s reverse transcription kit was used to construct cDNA libraries of the two tissues, oligo dT was used as primer for reverse transcription, and the cDNA ...
Embodiment 2
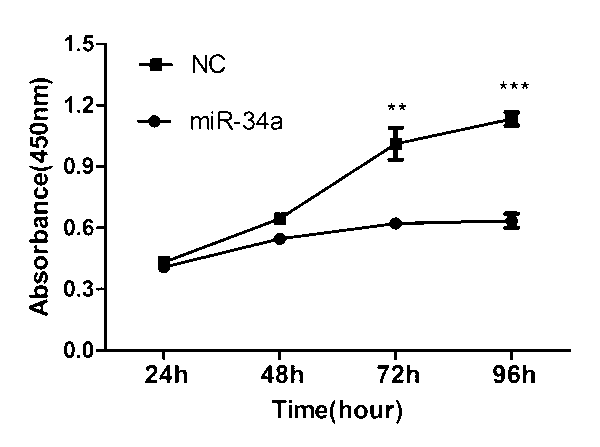
[0042] Example 2: The effect of miR-34a on the proliferation of H1299 cells
[0043] Spread the H1299 cells evenly in a 96-well plate with a medium of 1640. After 24 h, pass the mimics of miR-34a through lipo2000 transfection reagent and co-incubate with serum-free medium to transfer them into H1299 cells for transfection. After 6 h, replace the cell culture medium, put in fresh culture medium, then immediately add CCK8 reagent, incubate in the incubator for 2.5 h, then transfer the solution to the microplate, and detect the absorbance value at 450 nm every 24 h Measure once.
[0044] The test found that the transfection group had a significant decrease in absorbance compared with the control group, that is, the proliferation rate of the cells transfected with miR-34a was significantly lower than that of the control group, indicating that miR-34a can inhibit the proliferation of H1299 cells ( See figure 2 ).
Embodiment 3
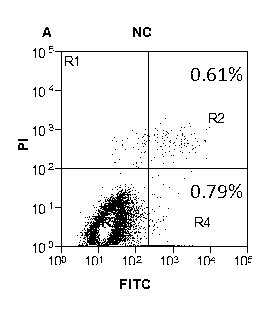
[0045] Example 3: The effect of miR-34a on H1299 cell apoptosis
[0046] Spread the H1299 cells evenly in a 6-well plate. After 24 hours, the miR-34a mimics were incubated with lipo2000 and serum-free medium 1640 and then transferred to H1299 cells. After 6 hours of transfection, the cell culture medium was replaced with fresh culture liquid. 48 h after transfection, the collected cells were stained with annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Sigma-Aldrich, USA). Flow cytometry was used to detect the apoptosis of H1299 cells. The results showed that the apoptosis rate of H1299 cells transfected with miR-34a was significantly higher than that of the control group (see image 3 , Figure 4 with Figure 5 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com