Separated polypeptide and application thereof
A technology of microorganisms and oligonucleotides, applied in the field of isolated polypeptides and their uses, can solve the problems of few aromatic amino acid transaminases and no reports, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Embodiment 1 Lactobacillus paracasei (Lactobacillus paracasei) M17 mutagenesis screening method
[0067] Pick an appropriate amount of bacteria (Lactobacillus sp. W2, preservation number CCTCC NO: M209318) from the activated slant and insert it into 5mL MRS liquid medium, culture it statically at 37°C for 12h, and inoculate the culture into Incubate in 100mL MRS liquid medium at 37°C for 18h. Take out 10mL culture medium, centrifuge at 8000rpm for 10min and wash twice with phosphate buffer, then add 0.1mol / L phosphate buffer with pH6.0 to adjust the cell concentration to 10 8 cfu / mL. Put the above-mentioned bacterial suspension in a plate with a diameter of 9 cm, irradiate under the ultraviolet lamp for 210 seconds, irradiate while stirring, take a sample in 5 mL of MRS liquid medium, incubate in the dark at 37°C for 1.5-2 hours, and then add the volume percentage of 0.6%, 0.8% and 1.0% diethyl sulfate solution, treated under dark light for 30-60 minutes, adding 2% so...
Embodiment 2
[0068] Cloning of embodiment 2 Lactobacillus paracasei M17 aromatic amino acid transaminase gene
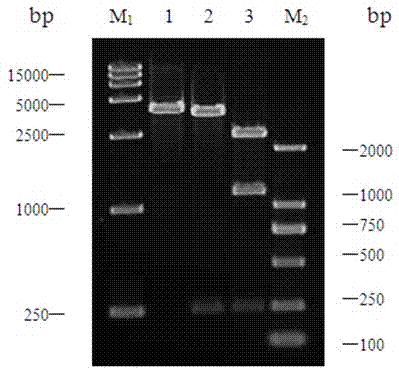
[0069] Pick Lactobacillus paracasei (Lactobacillus paracasei) M17 and inoculate it into 50mL MRS medium, culture it statically at 37°C for 14 hours, collect the bacteria by centrifugation, and extract Lactobacillus paracasei (Lactobacillus paracasei) M17 with TaKaRa company bacterial genome DNA extraction kit of genomic DNA. Primers were designed according to the Lactobacillus casei str. Zhang aromatic amino acid transaminase gene sequence in the NCBI database (Genbank accession number: NC_014334).
[0070] Upstream primer: 5'-GCGCC CATATG ACATTAGTTGATCGGATGAATC-3'
[0071] (SEQ ID NO: 3)
[0072] Downstream primer: 5'-CGC GTC GAC TTAATGCGTTGCCATATACTTGG-3' (SEQ ID NO: 4)
[0073] In order to facilitate cloning and ligation, an Nde I restriction site was designed at the 5' end of the upstream primer, and a Sal I restriction site was designed at the 5' end of the downstream...
Embodiment 3
[0075] Example 3 Construction of Lactobacillus paracasei M17 aromatic amino acid transaminase recombinant plasmid
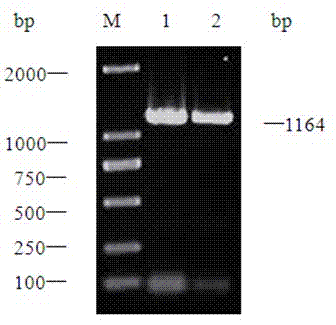
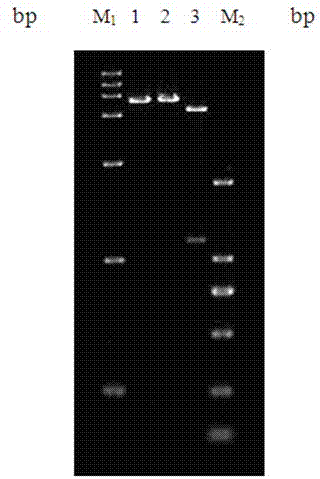
[0076] Plasmids pET-22b(+) and pMD19-ara T were extracted and digested with restriction endonucleases Nde I and Sal I for 3 hours. The target fragment was recovered by electrophoresis of the double-digested product, and ligated with the double-digested expression vector pET22b overnight at 16°C to form the recombinant plasmid pET22b-ara T. After verification by PCR and enzyme digestion, it was determined that the expression vector Lactobacillus paracasei transaminase recombinant plasmid was constructed successfully, as image 3 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com