L-lactic dehydrogenase derived from Lactobacillus casei and application of L-lactic dehydrogenase
A technology of lactate dehydrogenase and phenyllactic acid, applied in the field of L-lactate dehydrogenase, can solve the problems of low PLA concentration, pollution, and difficult separation of by-products
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] The cloning of embodiment 1 LcLDH1 gene and the construction of expression plasmid
[0027] According to the nucleotide sequence corresponding to L-LDH (CAP07851) of L. casei BL23, the following primers were designed:
[0028] LcLDH1-F: 5' -CATATG GTGGCAAGTATTACGGATAA-3', containing Nde I restriction site.
[0029] LcLDH1-R: 5'- CTCGAG CTGACGAGTTTCGATGTCATT-3', containing Xho I restriction site.
[0030] Extract the total DNA of Lactobacillus casei, use LcLDH1-F and LcLDH1-R as primers for PCR amplification, PCR conditions are: 94°C 4min; 94°C 30s, 51°C 30s, 72°C 1min 10s, 30 cycles; 72°C 10min. The PCR product was analyzed by 1% agarose gel electrophoresis, the target band was recovered by tapping the gel and ligated with the pUCm-T vector (pUCm-T-Lcldh1), transformed into E.coli JM109, and sent to Shanghai after the correct identification by bacterial liquid PCR The nucleotide sequence of LcLDH1 was obtained by biosequencing. The pUCm-T-Lcldh1 and pET-22b(+) p...
Embodiment 2
[0031] Example 2 Construction and Induced Expression of Recombinant Bacteria E.coli / Lcldh1
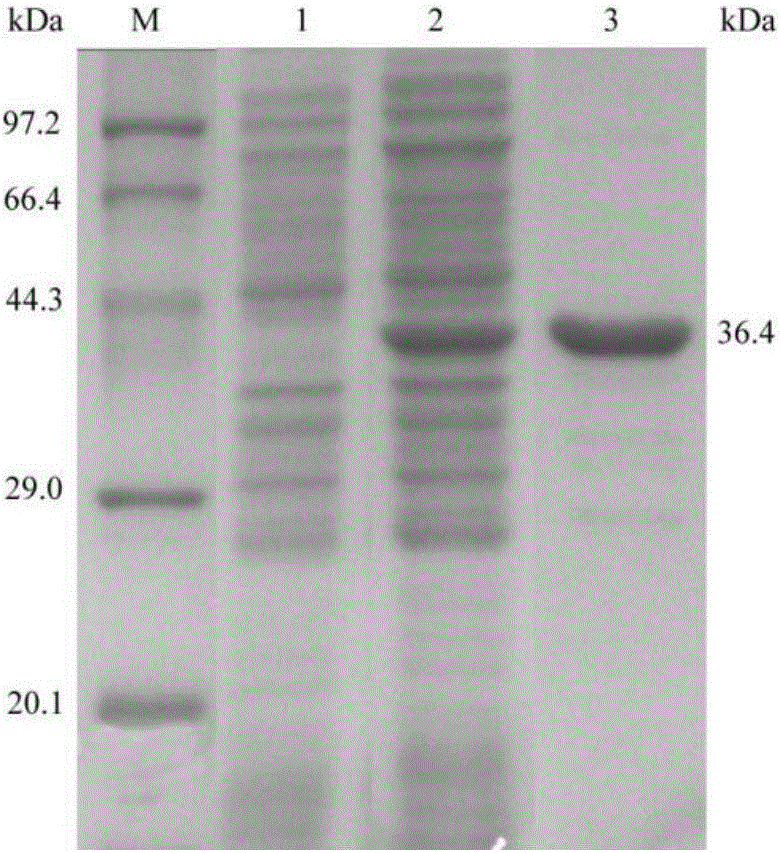
[0032] Transform pET-22b(+)-Lcldh1 into E.coli BL21(DE3), and after screening on a resistance plate containing Amp, pick positive transformants and place them in 2 mL of resistant LB liquid medium containing Amp, at 37°C 220rpm shaker shaking culture 14 ~ 16h. Transfer to 30mL of the same medium according to the inoculum size of 2%, and culture on a shaker at 220rpm at 37°C until the mid-logarithmic growth phase (OD600 =0.6~0.8), add IPTG to a final concentration of 0.4mmol / L, induce culture at 20°C and 220rpm for 8h. After the induction, the cells were collected by centrifugation and washed several times with phosphate buffer (pH=7.0) for SDS-PAGE analysis. Take the strain containing only empty pET-22b(+) as the control, see the results figure 1 , the recombinant strain E.coli / Lcldh1 (ie E.coli BL21(DE3)pET-22b(+)-Lcldh1) had a protein band at 36.4kDa with the same size as the targe...
Embodiment 3
[0033] Example 3 Whole cells of recombinant bacteria catalyze phenylpyruvate to generate L-PLA
[0034] Add 300 μL of bacterial suspension (30 mg of wet bacteria) and 250 μL of sodium phosphate buffer (pH 7.0) to a 1 mL centrifuge tube, incubate at 37°C for 5 min, then add 50 μL of glucose (final concentration 50 mmol / L) and 400 μL of phenylpyruvate ( Final concentration 10mmol / L), after reacting for 1h, take 200μL reaction solution and add 800μL methanol to terminate the reaction, pass through 0.22μm organic filter membrane, and carry out HPLC analysis; another 200μL reaction solution is extracted with 1mL ethyl acetate, and the upper organic phase is passed through After drying with anhydrous sodium sulfate, pass through a 0.22 μm organic filter membrane for enantioselective analysis. The results showed that after the system reacted for 40min, the concentration of L-PLA was 5.62mmol / L, and the productive rate was 56.2%, ee p >99%, after 80min of reaction, phenylpyruvate was...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Enzyme activity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com