Kit and method for detecting microRNA (ribonucleic acid)-122
A kit and asymmetric technology, applied in the field of kits for detecting microRNA-122
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
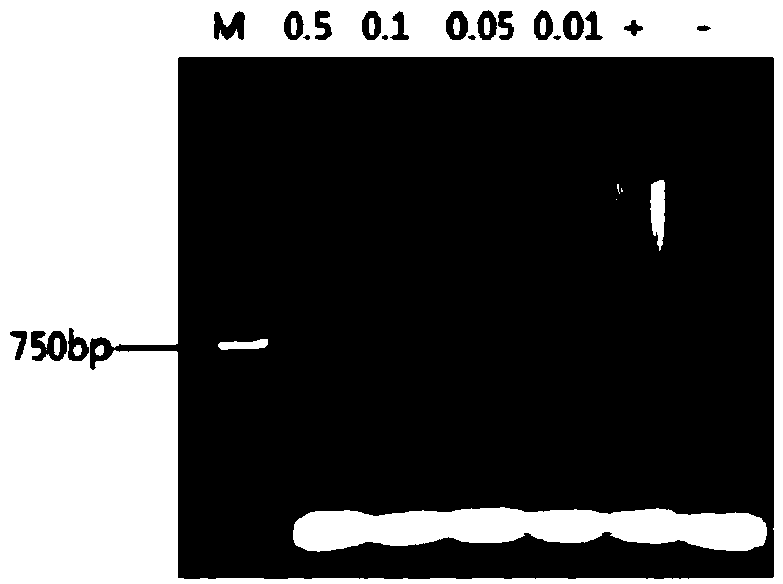
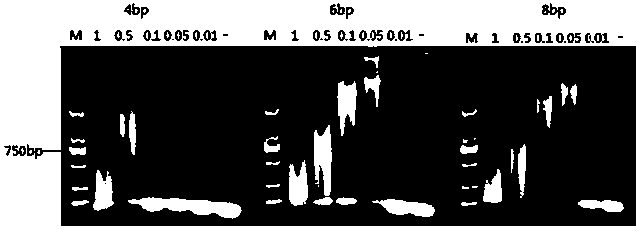
[0028] Embodiment 1, hairpin probe and kit for detecting microRNA-122
[0029] 1. Selection of detection target sequences and design of asymmetric hairpin probes
[0030] (1) Select the target sequence for detection according to the molecular biological characteristics of miRNA.
[0031] through literature or Genbank databases ( http: / / www.ncbi.nlm.nih.gov / ) to seize the human miRNA sequence related to cancer and determine it as the detection target sequence.
[0032] (2) Design of specific asymmetric hairpin probes
[0033] The Array Designer 4.0 software was used to design asymmetric hairpin probes for the target sequence, and the designed candidate probe sequences were submitted to the Genbank database for specificity analysis. Submit the designed asymmetric hairpin probes to The DINAMelt Web Server (http: / / mfold.rna.albany.edu / / ?q=DINAMelt / Hybrid2) for secondary structure analysis and between probes, between probes and The complementarity analysis of the target seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com