Aptamer EpCAM (epithelial cell adhesion molecule) B of EpCAM and preparation method thereof
An adhesion molecule and epithelial cell technology, applied in the field of nucleic acids, can solve the problems of poor antibody specificity, low binding force, and unsatisfactory clinical test results, and achieve the effects of small molecular weight, good permeability, and simple and rapid screening and detection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 In vitro screening of nucleic acid aptamers that specifically bind to epithelial cell adhesion molecule EpCAM
[0047] 1) Dissolve the synthesized 5nmol single-stranded DNA nucleic acid library in binding buffer (12mmol / L PBS, 0.55mmol / L MgCl 2 ), conduct heat treatment: heat at 95°C for 5 minutes, place on ice for 10 minutes, and then place at room temperature for 10 minutes;
[0048] 2) Incubate the processed single-stranded DNA nucleic acid library with Ni microbeads, and collect the liquid that is not bound to Ni microbeads;
[0049] 3) Incubate the liquid not bound to Ni microbeads with EpCAM Ni microbeads at 37°C for 40min;
[0050] 4) Wash the incubated EpCAM Ni microbeads with binding buffer, and then perform PCR reaction on the EpCAM Ni microbeads bound to oligonucleotides;
[0051] The PCR reaction program was: 94°C pre-denaturation for 3 minutes, 94°C for 30s, 53°C for 30s, 68°C for 30s, amplification for 10 cycles, and finally 68°C for 5 minutes....
Embodiment 2
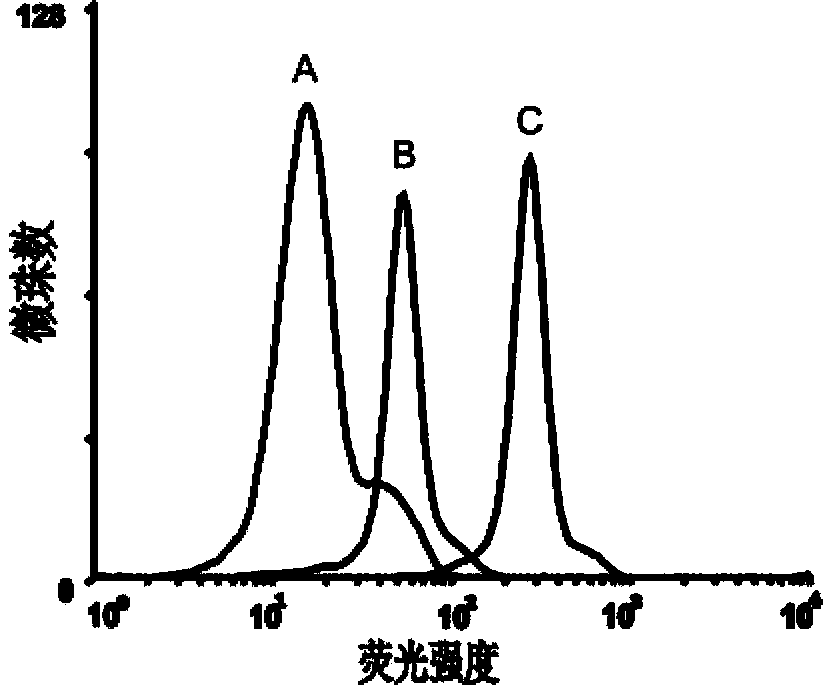
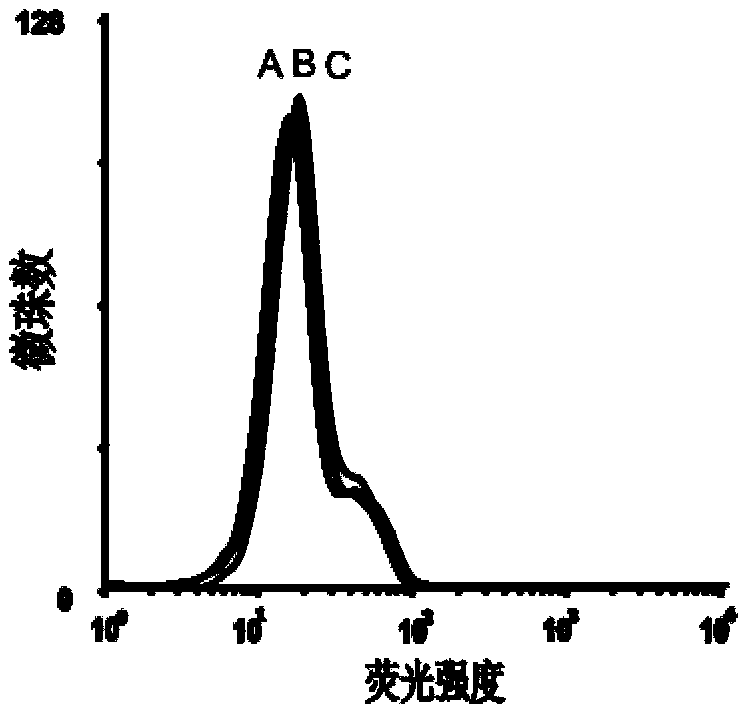
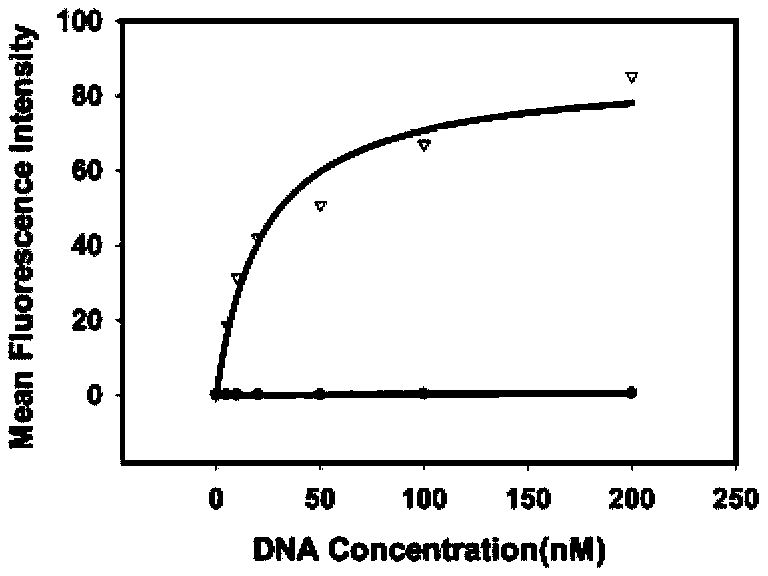
[0056] Example 2 Detection of the binding ability of the obtained single-stranded DNA to the epithelial cell adhesion molecule EpCAM by flow cytometry
[0057] First PCR amplifies the fluorescently labeled single-stranded DNA, using primer 2: 5'-Biotin-CTG ACC ACG AGC TCC ATT AG-3' and primer 3: 5'-FAM-AGC GTC GAA TAC CAC TAC AG-3' , the PCR product is double-stranded DNA with FAM at the 5' end and biotin at the 3' end, add streptavidin microbeads, react for 30min, and then use 0.1mol / L NaOH for single-stranded DNA, pass through a desalting column Purify to obtain FAM-labeled single-stranded DNA for flow analysis.
[0058] The dissociation constant (Kd=22.8 ±6.0). Use 200 μl of binding buffer to prepare DNA solutions of the above concentrations, heat at 95°C for 5 minutes, place on ice for 10 minutes, and then place at room temperature for 10 minutes. Add 155nmol / L EpCAM microbeads and incubate at 37°C for 40min. Wash the beads 3 times with binding buffer, then resuspend t...
Embodiment 3
[0060] Example 3 Screening to obtain nucleic acid aptamers that specifically bind to various cell lines
[0061] 1) Dissolve the synthesized 5nmol single-stranded DNA nucleic acid in the binding buffer (12mmol / L PBS, 0.55mmol / L MgCl 2 ), conduct heat treatment: heat at 95°C for 5 minutes, place on ice for 10 minutes, and then place at room temperature for 10 minutes;
[0062]2) Incubate the processed single-stranded DNA nucleic acid with 10,000 cells in a 24-well plate for 24 hours, at 37°C for 30 minutes or at 4°C for 40 minutes.
[0063] 3) After incubation, remove the buffer solution and wash twice, then scrape off the cells and dissolve them in 200 μL buffer solution, and use BD’s FACSAria flow cytometer to measure the fluorescence of the microbeads (see Figure 5 and 6 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com