Method for screening polychlorinated biphenyl degrading bacterium and polychlorinated biphenyl degrading bacterium
A technology for polychlorinated biphenyls and screening methods, applied in microorganism-based methods, biochemical equipment and methods, bacteria, etc., can solve the problem of microbial toxicity, low bioavailability of polychlorinated biphenyls, and difficulty in obtaining polychlorinated biphenyls. Benzene degrading bacteria and other problems, to achieve the effect of promoting degradation, simple method, and promoting complete degradation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
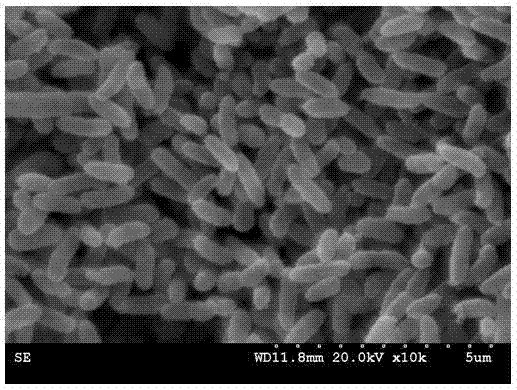
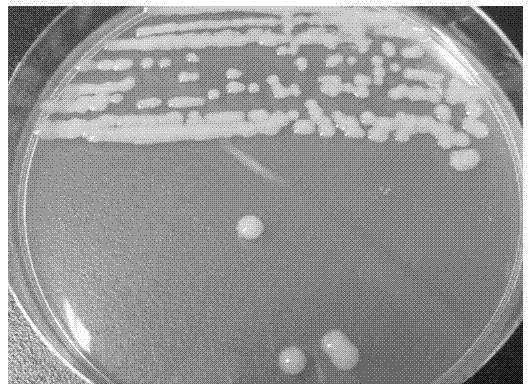
[0018] Example 1: Screening of PCB-degrading bacteria
[0019] The present invention screens out PCB-degrading bacteria from the environment through enrichment and domestication, separation and purification, and PCB degradation experiments. The specific method is as follows:
[0020] (1) Enrichment and domestication. The soil samples used to isolate microorganisms were taken from a PCB-contaminated area in Luqiao District, Taizhou City, Zhejiang Province. Inorganic salt medium including KH 2 PO 4 1.00g / L, K 2 HPO 4 ·3H 2 O3.00g / L, MgSO 4 0.15g / L, FeSO 4 0.01g / L, CaCl 2 0.005g / L, NaCl1.00g / L, (NH 4 ) 2 SO 4 0.50g / L, trace element solution 0.1% (v / v), pH7.2; trace element solution formula is Na 2 Mo 4 ·H 2 O6.70g / L, ZnSO 4 ·5H 2 O28.00g / L, CuSO 4 ·5H 2 O2.00g / L, H 3 BO 4 4.00g / L, MnSO 4 ·5H 2 O4.00g / L, CoSO 4 ·7H 2 O4.70g / L, pH7.2. Fill 100 mL of the above culture medium into a 300 mL Erlenmeyer flask, and sterilize at 121 °C for 20 min before use. Befo...
Embodiment 2
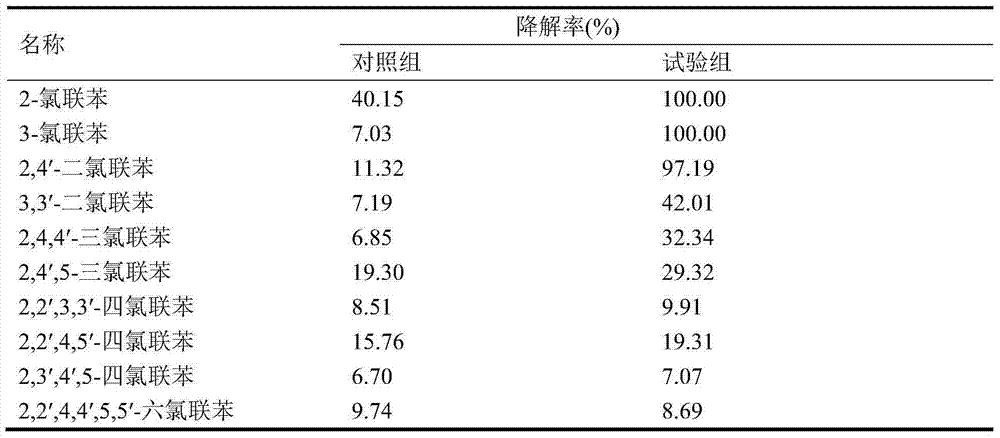
[0027] Embodiment 2: Degradation of bacterial strain HC3 to benzoic acid and chlorobenzoic acid
[0028] Degradation of benzoic acid and chlorobenzoic acid. The degradation system is the same as in Example 1 (4), and the degradation substrates are benzoic acid, 2-chlorobenzoic acid, 3-chlorobenzoic acid and 4-chlorobenzoic acid, and the concentration of each substrate is 20-30 mg / L. After incubating at 180r / min and 30°C in the dark for 3 days, add 3mol / L HCl to each tube to adjust the pH to 2.5, then add 15mL ethyl acetate and seal the tube opening. Vortex for 5 minutes, sonicate for 15 minutes, centrifuge at 2500r / min for 5 minutes, take the upper organic phase and dry it with nitrogen, redissolve the sample in methanol, and finally transfer it to a sample vial for HPLC analysis. The high-performance liquid chromatograph is Agilent1100, and the DAD detector is selected, and the chromatographic column is Zorbax Eclipse XDB-C18 (4.6mm×250mm, 5μm). The ratio of mobile phase me...
Embodiment 3
[0032] Example 3: Degradation of polychlorinated biphenyls in soil by bacterial strain HC3
[0033] The soil (with a PCB content of 18.60 mg / kg) taken from a PCB-contaminated site in Taizhou City, Zhejiang Province was air-dried naturally, ground finely, passed through a 20-mesh sieve, mixed evenly, and divided into 200 g in each beaker. Then inoculate the bacterial strain HC3 (the preparation method of the bacterial solution is the same as the method described in Example 1) so that the bacterial concentration in the soil reaches 1.0×10 8 cfu / g (the same amount of heat sterilization was added to the control group), and an appropriate amount of water was added to make the soil moisture content reach 40%. The test group was placed in an artificial climate room, protected from light, and the soil temperature was maintained at 22-23°C during the test. On the fifth and tenth day of the test, soil samples were taken from each test group by the five-point method for the determinatio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com