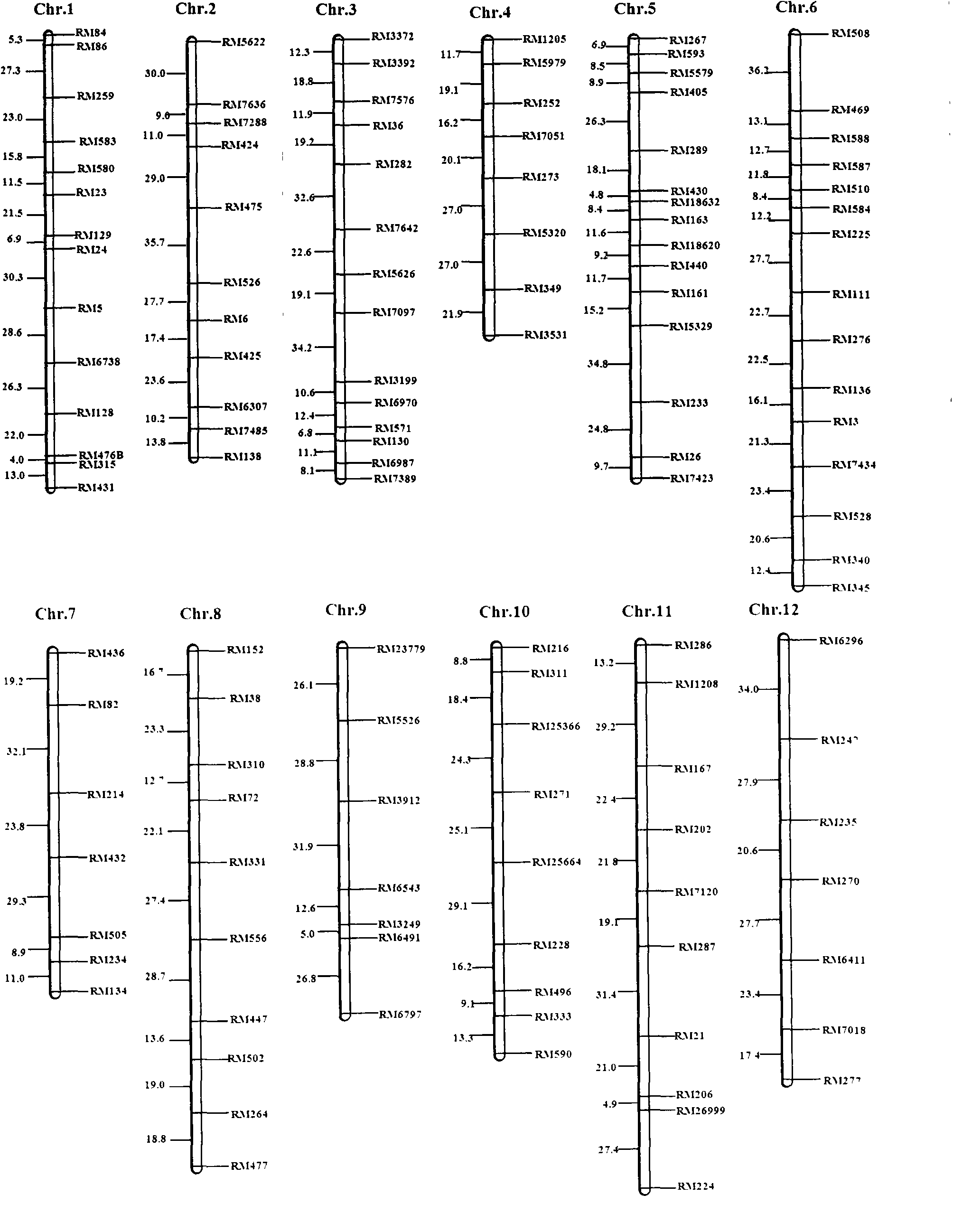
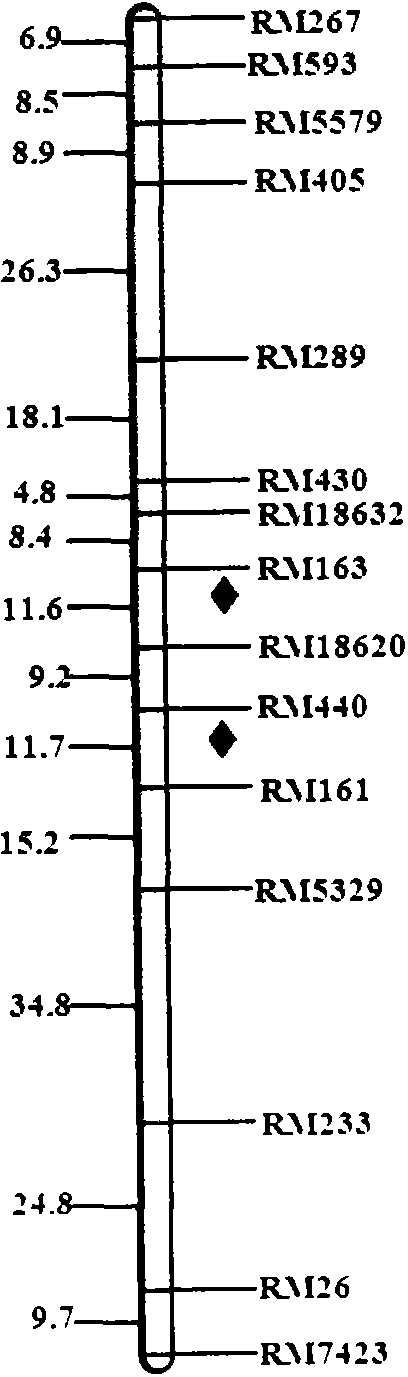
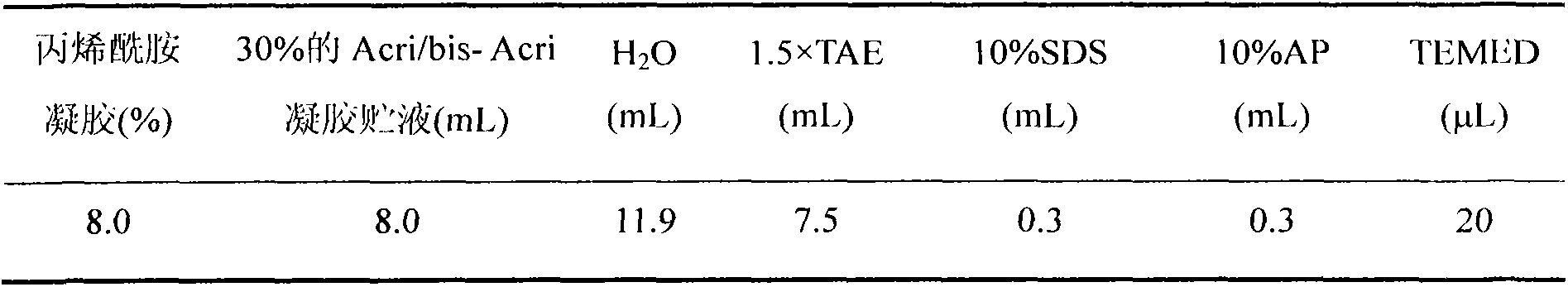SSR (simple sequence repeat) markers, linked with LRPW (loss rate of panicle weight) related Aphelenchoides besseyi Christie resistant QTL (quantitative trait loci), on chromosome 5 and application thereof
A technology for resistance to A. rice nematode, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of limiting the effective use of resources for resistance to A. rice, and improve breeding efficiency , speed up the breeding process, simplify the effect of selection methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1, Acquisition of Molecular Markers Linked to the Resistance QTL of D.
[0020] 1. Plant material
[0021] In 2008, Huaidao 5 and Tetep were planted in the field of Jiangsu Academy of Agricultural Sciences and crossed to obtain the hybrid F 1 , next year F 1 F 2 Seeds, propagated F in Hainan in 2009 2 group, as a mapping group. f 2 Individual plant number, parent and hybrid F at tillering stage 1 and F 2 Some leaves of each individual plant in the population were stored in a -70°C refrigerator for SSR analysis, F 2 Harvest the individual plants of the population for phenotypic identification.
[0022] 2. Cultivation and isolation of nematodes
[0023] Inoculate Botrytis cinerea (Botrytis cinerea) bacterial block on Potato Dextrose Agar (PDA) medium and cultivate at 25°C. After Botrytis cinerea overgrows the medium, use 3% hydrogen peroxide to sterilize the surface of rice dry acerbone nematode for 10min , after washing with sterilized ultrapure water f...
Embodiment 2
[0060] Example 2, the application of molecular markers on chromosome 5 linked to the resistance QTL of rice stem nematode resistance to rice 5 and Tetep in the offspring of the cross combination with Huaidao 5 and Tetep as parents
[0061] The SSR marker obtained on chromosome 5 linked to the rice stem nematode resistance QTL, the F 2∶3 Part of the individual plants of the family were predicted for the resistance of rice stem nematode, and the DNA of each individual plant was extracted, and then PCR amplification analysis was performed with the primers of SSR markers RM163, RM18620, RM440 and RM161, and the band type analysis was used to determine whether there were corresponding The presence of the marker indicates that the line has reached the level of resistance to A. sativa, and the absence of the marker indicates that it is susceptible to the disease. Subsequently, the actual resistance of the tested lines to rice A. spp. was determined by using the artificial inoculation...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com