Method for identifying sugarcane germplasm resources based on SSR (Simple Sequence Repeats) and CE (capillary electrophoresis) technique
An identification method and sugarcane technology, applied in the field of high-throughput detection, can solve problems such as difficulty in identification of sugarcane germplasm resources, and achieve the effects of reducing workload, accurate experimental results, and reducing human errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
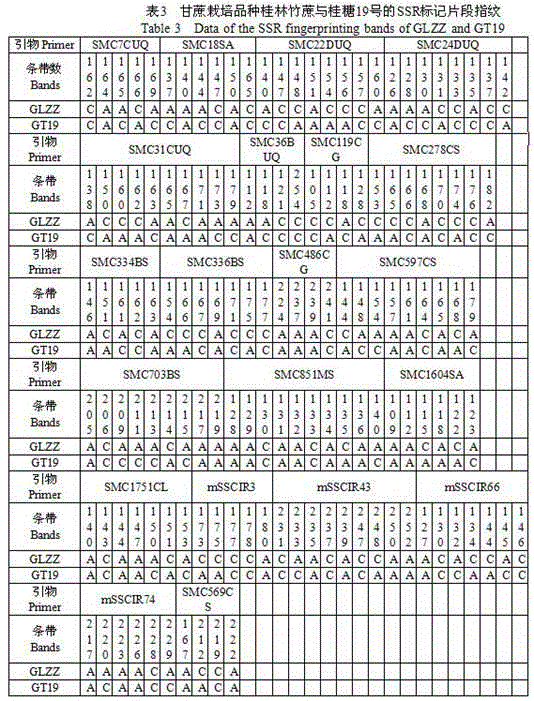
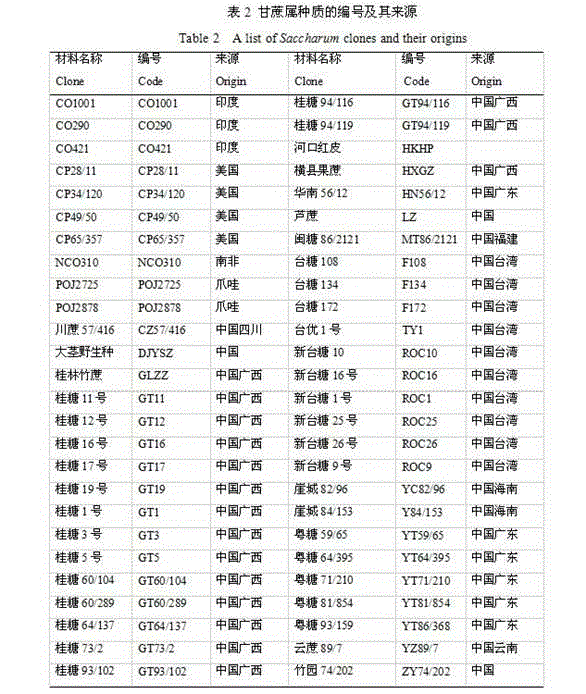
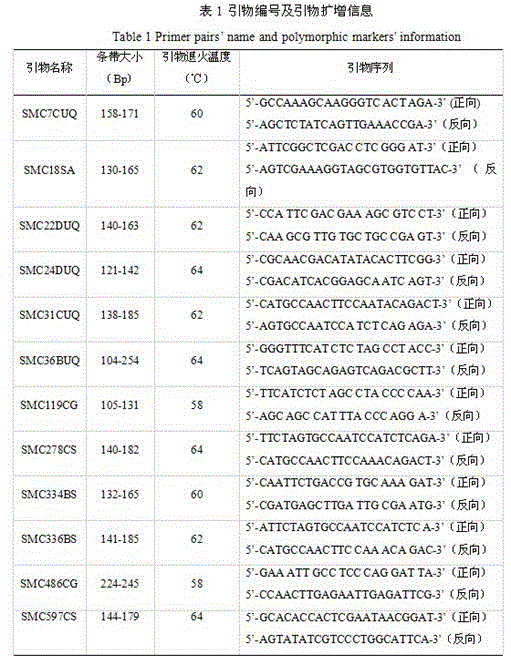
[0015] Example 1: 12 pairs of SSR fluorescent primers were used to establish plant maps of 52 sugarcane cultivars, and the selected 52 sugarcane cultivars are shown in the table below.
[0016]
[0017] 1. Genomic DNA extraction
[0018] (1) Take young sugarcane leaves in a mortar, add liquid nitrogen to grind them into powder, and put them into a 2 ml centrifuge tube;
[0019] (2) Add 1.2 ml 65°C preheated SDS extraction solution (Tris-HCl 200 mmol / L, pH8.0; EDTA 50 mmol / L pH8.0; NaCl 500 mmol / L; 3% SDS) into the centrifuge tube , Shake the sample powder and the extract quickly to prevent them from forming agglomerates, put in a water bath at 65°C for 45 minutes, and shake gently 4-6 times during the period;
[0020] (3) Add 200 μl KAc (5 mol / L, pH 4.8), mix well, and place in ice bath for 15 min;
[0021] (4) Centrifuge at 10,000 rpm for 15 min in a refrigerated centrifuge at 4°C;
[0022] (5) Take 800 μl supernatant, add 800 μl chloroform to mix, and centrifuge at 120...
Embodiment 2
[0040] Embodiment 2: the identification of sugarcane variety to be tested
[0041] 1. Select two kinds of unknown sugarcane varieties as materials to be tested, and carry out DNA extraction, SSR-PCR amplification, capillary gel electrophoresis detection and result data analysis according to the method in Example 1;
[0042] 2. According to the read band size, according to the 141 specific SSR marker fragments in Example 1, if it contains these 141 specific SSR marker fragments, it will be marked as "A", if there is no extension, it will be marked as "C". , to construct the SSR fingerprint of the sugarcane variety to be tested. And comparing the results with the established standard sugarcane fingerprints in Example 1, the sugarcane variety was identified as Guitang No. 19.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com