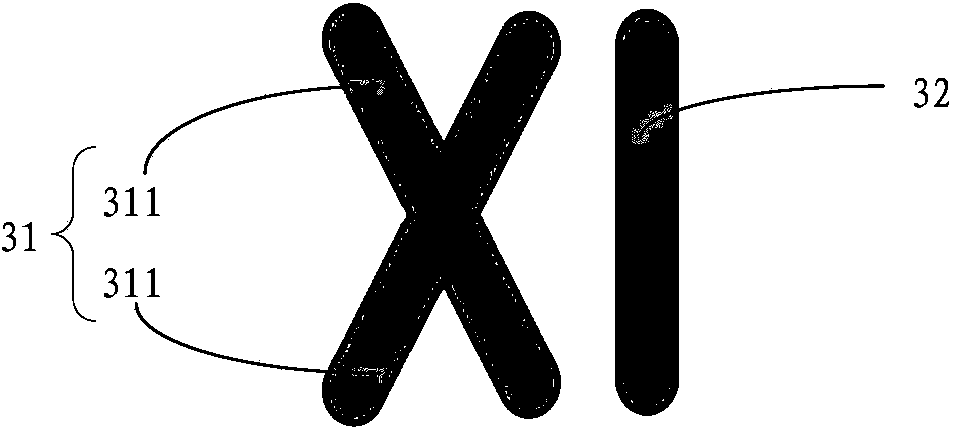
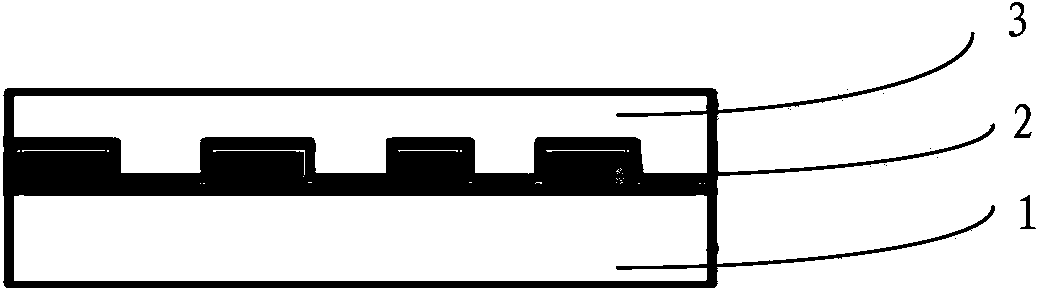
LSPR (localized surface plasma resonance) sensing device and preparation method thereof as well as DNA detection method
A technology of a sensing device and a manufacturing method, applied in the field of biochemical detection, can solve the problems of single incident light and lack of sensitivity, achieve the effects of accurate detection, avoid wave peak broadening, and improve accuracy and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0089] In the present invention, select Gram-positive bacteria to carry out the present invention's operation as bacterial standard sample:
[0090] S11, take 0.5-2ml of the cultured bacteria solution in the EP tube, centrifuge at high speed for 30s, and discard the supernatant; add buffer RB to the EP tube to resuspend, and discard the supernatant after high-speed centrifugation; repeat this step 2 to 3 times;
[0091] S12, add an appropriate amount of lysozyme, mix upside down, and incubate at 37°C for 30-60 minutes; after high-speed centrifugation, discard the supernatant and resuspend the cells in buffer RB by shaking or pipetting;
[0092] S13, add 20 μl of RNase A (25 mg / ml) solution to the solution obtained in step S13, shake and mix, and place at room temperature for 5-10 minutes;
[0093] S14, add the binding solution CB, and then add a small amount of isopropanol, at this time there will be flocculent sediment, immediately vortex and mix thoroughly to obtain a mixtur...
Embodiment 2
[0122] Based on the above comparison, in Example 2, the DNA extracted from Staphylococcus aureus is used as the DNA to be tested to carry out the steps in Example 1, and the DNA capture probe still uses the 16srRNA of Gram-positive bacteria in Example 1 The capture probe sequence of the gene; the detection step is carried out according to the same steps as in Example 1.
[0123] In the final absorption spectrum, its absorption peak obtained does not move, then it is because the probes fixed on the surface of the DNA to be measured and the LSPR sensor (that is, the quartz glass plate of the above-mentioned asymmetrical XI nano-metal structure) cannot be combined, That is to say, the detected bacteria are not the bacteria corresponding to the probe.
[0124] Of course, in the repeated implementation of the above embodiment, there may be a result that the absorption peak moves to the left, because it indicates that the fixed probe falls off, causing the absorption peak to move to t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com