Engineering bacteria and method for preparing tert-butyl (3R,5S)-6-chloro-3,5-dihydroxyhexanoate
A technology of engineering bacteria, pet-30a, applied in the direction of microorganism-based methods, biochemical equipment and methods, bacteria, etc., can solve the problems of increasing costs, unrealizable, increasing process flow, etc., to reduce production costs and avoid additives Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1 Construction of plasmid pET30-AdhR
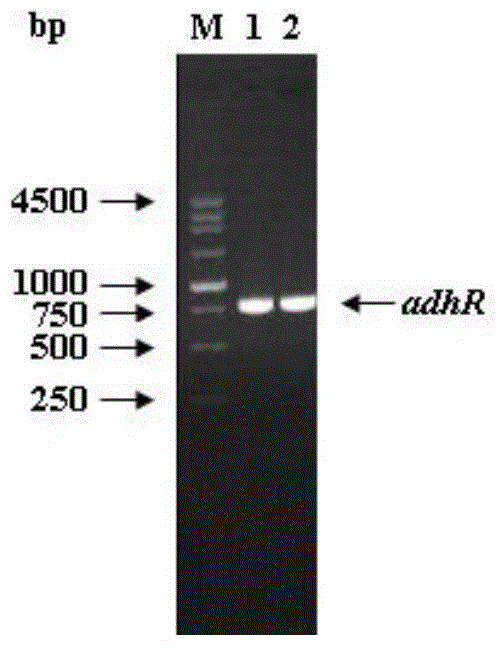
[0064] The adhR gene of the Lactobacillus kefir DSM20587 bacterium was cloned using primers F_AdhR / R_AdhR to obtain an adhR gene with a length of 759bp, and the gene size was verified by nucleic acid electrophoresis, such as figure 1 .
[0065] The sequence of primer F_AdhR is:
[0066] 5'-CATGCCATGGCTATGACTGATCGTTTAAAAG-3';
[0067] The sequence of primer R_AdhR is:
[0068] 5'-CCCGAGCTCTTATTGAGCAGTGTATCCAC-3'.
[0069] The NCBI accession number of the adhR gene sequence is AY267012.
[0070] The adhR gene was digested with NcoI and SacI, and the gene band after digestion was recovered. At the same time, the pET-30a(+) plasmid was digested with NcoI and SacI, the plasmid band after digestion was recovered, and the digested adhR The gene and digested pET-30a(+) plasmid were ligated with ligase and transformed into the cloning host Escherichia coli DH5α. Colony PCR was performed with primers F_AdhR / R_AdhR to verify the...
Embodiment 2
[0071] Example 2 Construction of plasmid pACYCDuet-CB
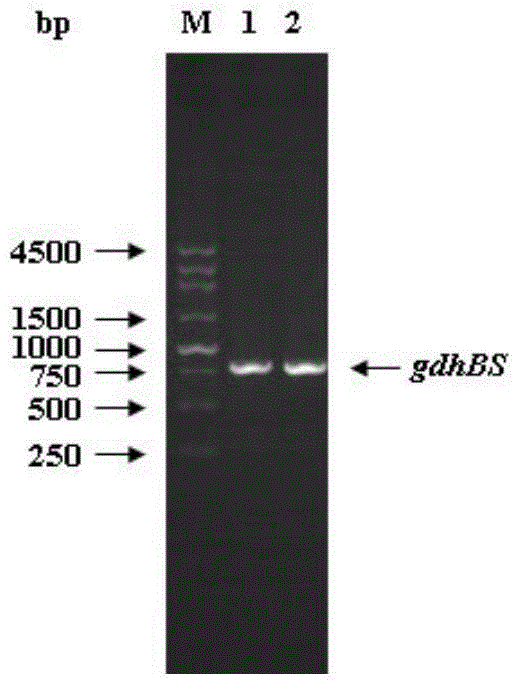
[0072] The gdhBS gene of Bacillus subtilis168 bacteria was cloned using primers F_GdhBS / R_GdhBS to obtain a gdhBS gene with a length of 786bp, and the gene size was verified by nucleic acid electrophoresis, such as figure 2 .
[0073] The sequence of primer F_GdhBS is:
[0074] 5'-GAAGATCTCATGTATCCGGATTTAAAAGGAAAAGTC-3';
[0075] The sequence of primer R_GdhBS is:
[0076] 5'-CCGCTCGAGTTAACCGCGGCCTG-3'.
[0077] NCBI accession number of gdhBS gene sequence: AL009126.
[0078] The gdhBS gene was digested with BglII and XhoI, and the gene band after digestion was recovered. At the same time, the pACYCDuet-1 plasmid was digested with BglII and XhoI, and the plasmid band after digestion was recovered. The digested gdhBS gene and enzyme The cut pACYCDuet-1 plasmid was ligated with ligase and transformed into the cloning host Escherichia coli DH5α. Colony PCR was performed with primers F_GdhBS / R_GdhBS to verify the trans...
Embodiment 3
[0088] Example 3: Construction and induced expression of genetically engineered bacteria EcoR
[0089] The plasmid pET30-AdhR constructed in Example 1 was transformed into the expression host Escherichia coliBL21 (DE3), and colony PCR was performed with primers F_AdhR / R_AdhR to verify the transformed recombinant. The verified genetically engineered bacteria is EcoR. The EcoR was inoculated into the fermentation medium and cultured with constant temperature shaking for 12-16 hours. The culture conditions were 37° C. and 200 rpm. When the cell concentration grows to OD 600 When =1.0, add 0.6mM IPTG (final concentration) and induce at 16°C for 20h.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com