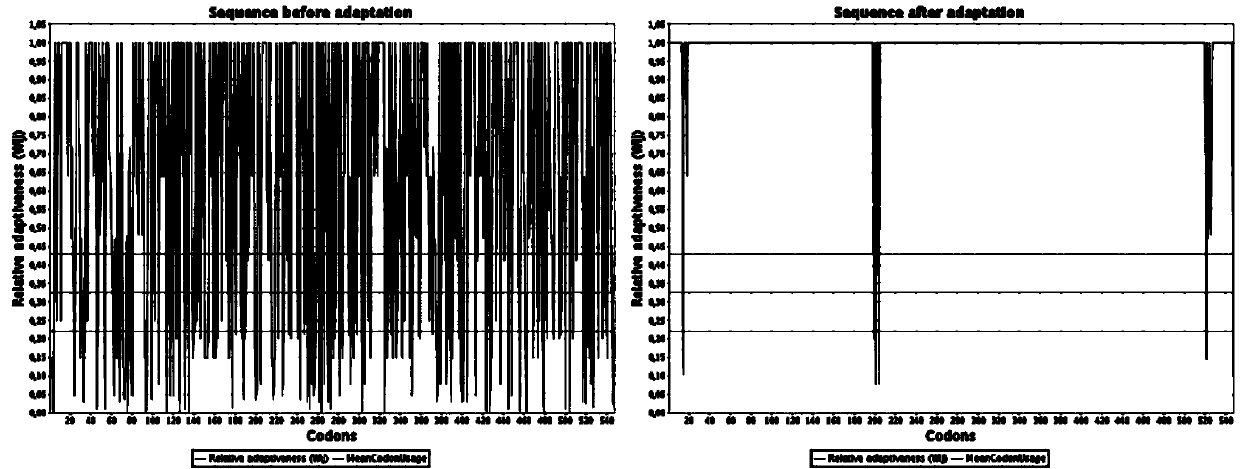
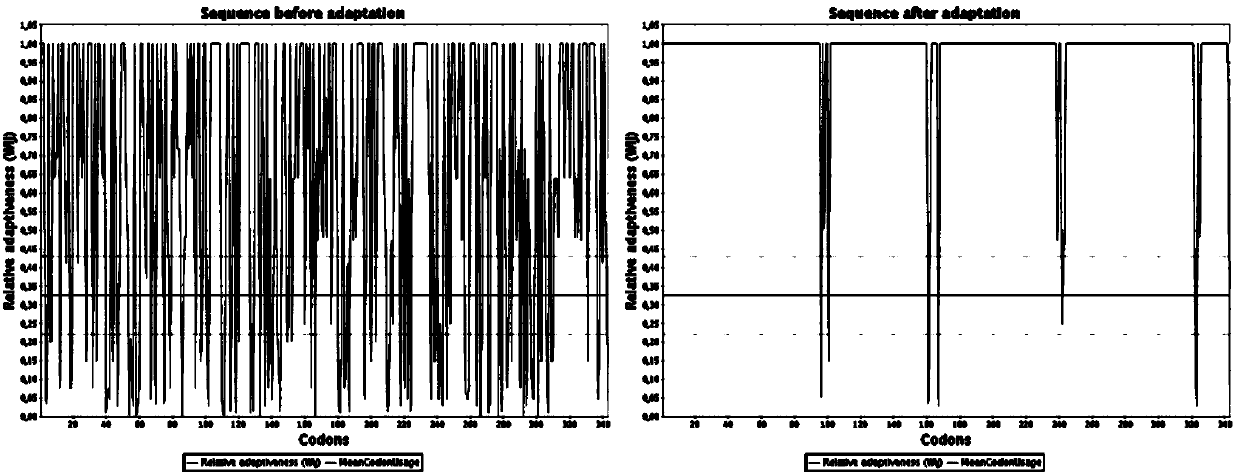
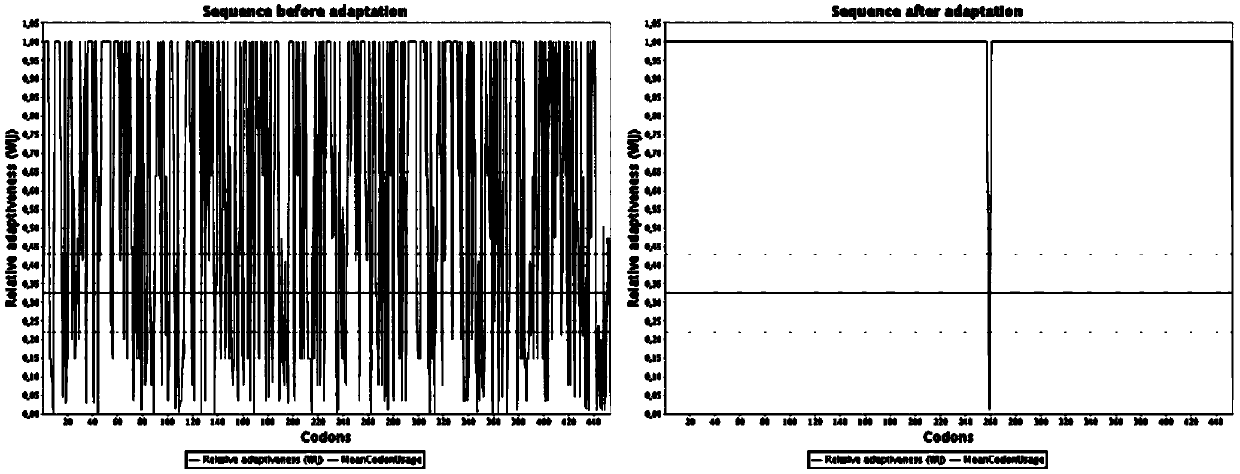
Engineering bacterium based on cellulose metabolic pathway key enzyme and implementation method of engineering bacterium based on cellulose metabolic pathway key enzyme
A technology of engineering bacteria and cellulose, applied in the field of engineering bacteria of β-1,4-glucosidase, can solve the problem of low expression level, and achieve the effect of ensuring quality, improving protein activity and solubility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] In this example, Streptomyces griseorubens was isolated from rotten bacteria collected in Pujiang Town, Shanghai.
[0031] Straw, the preservation number is CGMCC No.5706. The strain was inoculated in LB liquid medium and cultured at 32°C for 48 h.
[0032] The above LB liquid medium components are: peptone 10.0g / L, yeast extract 5.0g / L, NaCl 10.0g / L, pH 6.8-7.2. Add 15.0‐20.0g / L agar to the liquid medium to obtain LB solid medium.
[0033]1) Genomic DNA extraction of Streptomyces griseorubens: collect 2.0 mL of bacterial liquid, and centrifuge at 12000 rpm for 2 min. Discard the supernatant, collect the bacterial pellet, add 180μL lysozyme (20mg / mL) and 20μL EDTA solution (0.5M, pH8.0), treat at 37℃ for 45min, add 4μL RNase A (100mg / mL), shake and mix for 15s , placed at room temperature for 5 minutes, and then completed the remaining operations according to the instructions of the bacterial DNA extraction kit (TIANGEN) to obtain high-purity genomic DNA. The qualit...
Embodiment 2
[0038] Construction of co-expression vector
[0039] 1) According to the optimized nucleotide sequence, design the PCR primer sequence containing restriction site as follows:
[0040] EX‐Nco I‐F: TACCATGGCTGCTGTTCCGTGCACCGTTGACTACA
[0041] EX‐BamH I‐R: CGGGATCCTTAATGATGATGATGATGATGAGAAACCGGCGGGTAAGCG
[0042] EN‐Nde I‐F:TACATATGGACACCACCCCTGTGCGAAGAATTCGGT
[0043] EN‐Xho I‐R: CCCTCGAGTTAATGATGATGATGATGATGAGCGGTACGGCAAGCGGTA
[0044] BG‐Msc I‐F:GATGGCCATGGTGACCATCGACTACGCTGCTCTGC
[0045] BG‐Xho I‐R: CCCTCGAGTTAATGATGATGATGATGATGAGCAGCTTCACGAACACGT
[0046] 2) Using pUC57-EX as a template, PCR amplification was performed with primers containing Nco I and BamH I restriction sites to obtain the exo‐β‐1,4‐glucanase gene sequence, using DNA A‐Tailing Add A to Kit and connect to T‐Vector PMD TM 19‐T (TaKaRa), and the ligation product was transferred into DH5α E. coli. Positive clones were selected, and plasmids were extracted by shaking bacteria for sequencing. If correct,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com