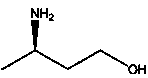
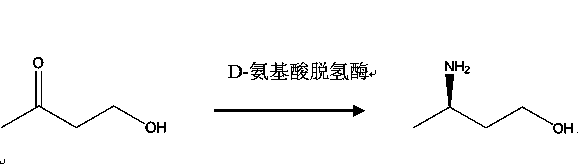
Method for producing R-3-aminobutanol
A technology of aminobutanol and its production method, which is applied in the direction of biochemical equipment and methods, enzymes, and the use of carriers to introduce foreign genetic material, etc., can solve the problems of no biocatalysis, etc., and achieve the effects of easy separation, reduced separation cost, and low price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0017] A kind of production method of R-3-aminobutanol, its preparation method comprises the following steps:
[0018] Step 1, preparation of recombinant D-amino acid dehydrogenase genetically engineered bacteria
[0019] Choose from Corynebacterium glutamicum The D-amino acid dehydrogenase gene sequence of ATCC13032 was artificially designed, and the designed gene sequence is shown in SEQ ID NO: 1; the sequence was cloned into the expression vector pET22b by Nde I and Hind III enzymes through whole gene synthesis site, transform Escherichia coli DH5α competent cells; pick positive transformants and sequence and identify them, and obtain recombinant expression vectors; transfer recombinant expression vectors into Escherichia coli BL21 (DE3) strains, and obtain recombinant D-amino acid desensitizers that can induce expression Hydrogenase recombinant D-amino acid dehydrogenase genetically engineered bacteria;
[0020] Step 2, preparation of recombinant D-amino acid dehydroge...
Embodiment 1
[0026] A kind of production method of R-3-aminobutanol, its preparation method comprises the following steps:
[0027] Step 1, preparation of recombinant D-amino acid dehydrogenase genetically engineered bacteria
[0028] Choose from Corynebacterium glutamicum The D-amino acid dehydrogenase gene sequence of ATCC13032 was artificially designed, and the designed gene sequence is shown in SEQ ID NO: 1; the sequence was cloned into the expression vector pET22b by Nde I and Hind III enzymes through whole gene synthesis site, transform Escherichia coli DH5α competent cells; pick positive transformants and sequence and identify them to obtain recombinant expression vectors; transfer the recombinant expression vectors into Escherichia coli BL21 (DE3) strains to obtain recombinant D-amino acid desensitizers that can induce expression Hydrogenase recombinant D-amino acid dehydrogenase genetically engineered bacteria;
[0029] Step 2, preparation of recombinant D-amino acid dehydroge...
Embodiment 2
[0035] A kind of production method of R-3-aminobutanol, its preparation method comprises the following steps:
[0036] Step 1, preparation of recombinant D-amino acid dehydrogenase genetically engineered bacteria
[0037] Choose from Corynebacterium glutamicum The D-amino acid dehydrogenase gene sequence of ATCC13032 was artificially designed, and the designed gene sequence is shown in SEQ ID NO: 1; the sequence was cloned into the expression vector pET22b by Nde I and Hind III enzymes through whole gene synthesis site, transform Escherichia coli DH5α competent cells; pick positive transformants and sequence and identify them, and obtain recombinant expression vectors; transfer recombinant expression vectors into Escherichia coli BL21 (DE3) strains, and obtain recombinant D-amino acid desensitizers that can induce expression Hydrogenase recombinant D-amino acid dehydrogenase genetically engineered bacteria;
[0038] Step 2, preparation of recombinant D-amino acid dehydroge...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com