Method for analyzing protein O-glycosylation sites
A technology for glycosylation sites and proteins, which is applied in the field of analyzing protein O-glycosylation sites, can solve the problems of undiscovered, difficult to control chemical methods, and difficult to analyze O-glycosylation sites, and achieves a simple method. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
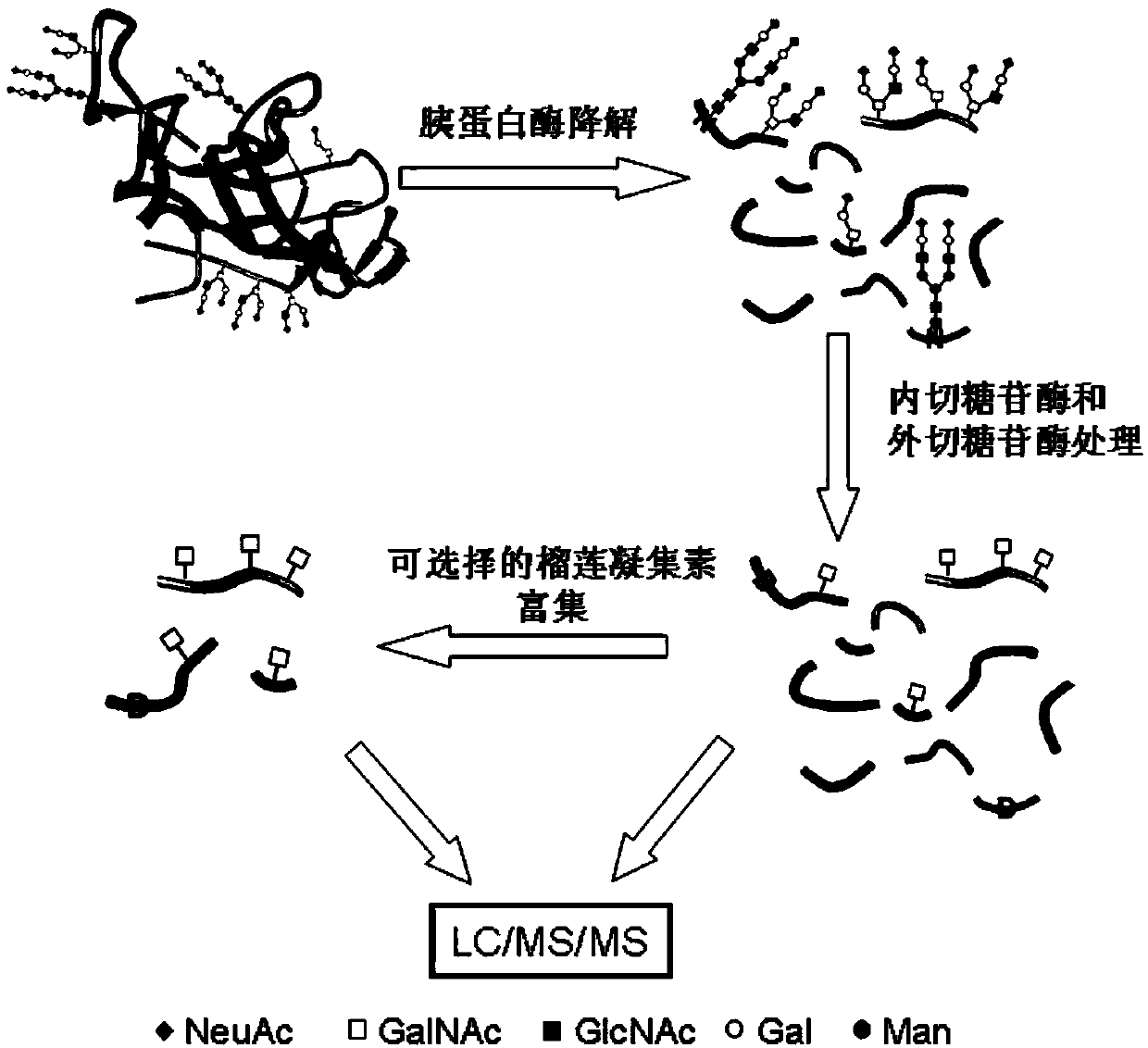
[0007] A method for analyzing protein O-glycosylation sites, the steps are as follows:
[0008] (1) After the proteome sample or a single protein sample is cleaved with endonuclease, the N-glycosidase and exoglycosidase are combined to cut off the N-sugar chain and part of the O-sugar chain, and dried under reduced pressure to obtain the Omics protein samples or single protein samples of GalNAc sugar-tagged peptides and non-glycopeptides;
[0009] (2) Load the omics protein samples with GalNAc sugar tags and non-glycopeptides prepared in step (1) on the durian lectin column for separation, and collect the enzyme-digested peptides with GalNAc sugar tags The peptide solution is dried under reduced pressure to prepare the hismic protein sample;
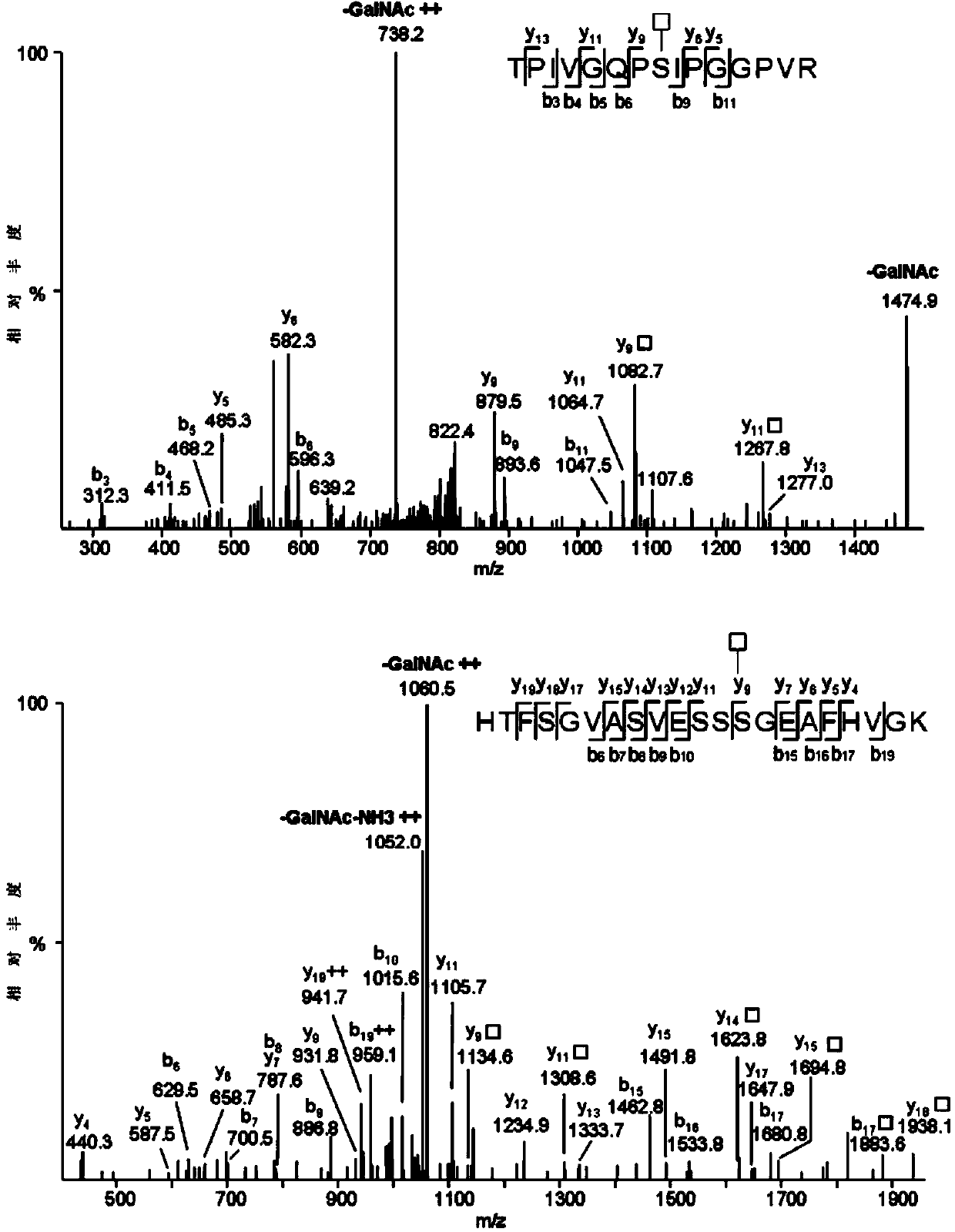
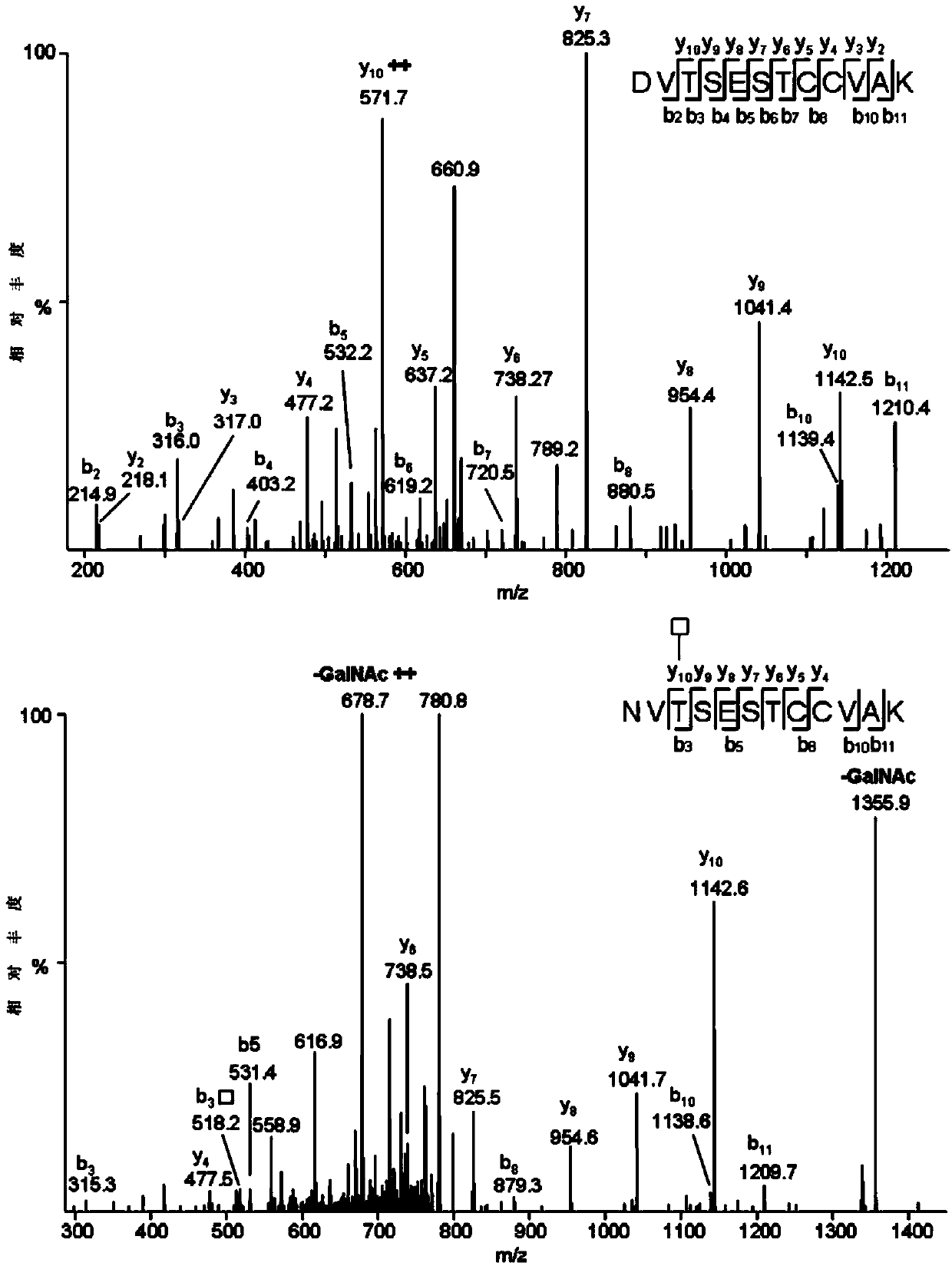
[0010] (3) Use the single protein sample prepared in step (1) or the hismic protein sample prepared in step (2) to 18 Separation by reversed-phase chromatographic column, and then detection by high-resolution mass spectrometer in posit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com