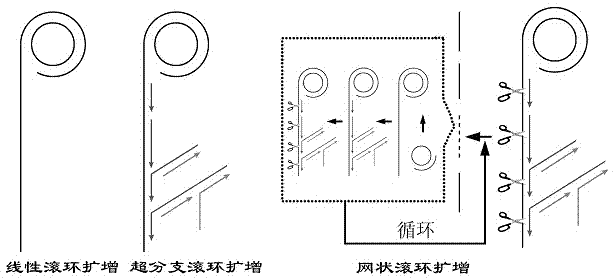
Nicking endonuclease-based netted rolling cycle amplification system and use thereof
A technology of nicking endonuclease and rolling circle amplification, which is applied in the field of network rolling circle amplification system based on nicking endonuclease
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
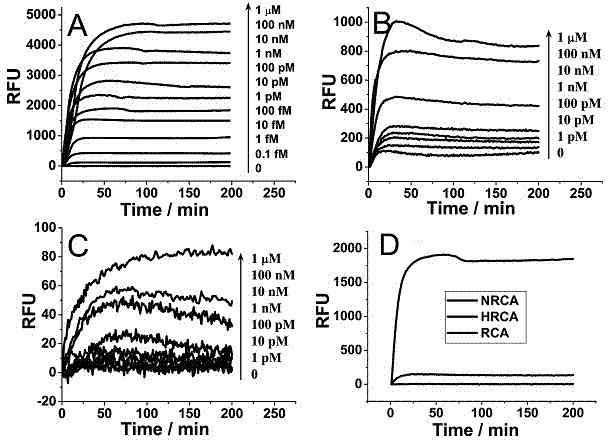
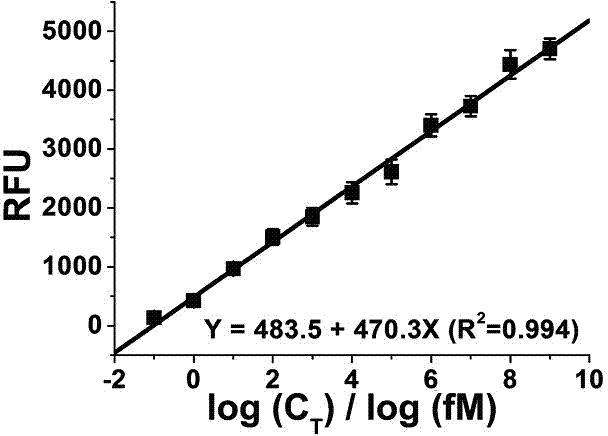
[0032] Example 1: Electrophoretic characterization of target DNA amplification products with different concentrations
[0033] 100 nM circular DNA template, different concentrations of target DNA, 1 μM primer, 400 μM dNTP, 8 U Bst DNA polymerase large fragment, 10 U nicking endonuclease Nb. BsrDI, 1×Bst DNA polymerase large fragment buffer solution (20 mM Tris-HCl, 10 mM KCl, 10 mM (NH 4 ) 2 SO 4 , 2 mM MgSO 4 , 0.1% Triton X-100, pH 8.8) were mixed, and double-distilled water was added to make the total volume of the reaction system reach 50 μL. After the reaction was placed in a metal bath at 65 °C for 1 hour, the reaction was terminated at 95 °C for 10 minutes. The main difference from the existing hyperbranched rolling circle amplification reaction system is the use of the nicking endonuclease Nb.BsrDI. Mix 5 μL sample and loading buffer solution 6×loading buffer (1 μL) evenly, and pour into the sample well. The voltage was set to 80 V and the time was 30 min for agar...
Embodiment 2
[0035] Example 2: Electrophoretic characterization of target DNA amplification specificity
[0036] 100 nM circular DNA template, 1 μM target DNA, DNA with 1, 3, 5 base mismatches with the target DNA, and DNA completely irrelevant to the target DNA, 1 μM primer, 400 μM dNTP, 8 U of Bst DNA polymerase large fragment, 10 U of nicking endonuclease Nb. BsrDI, 1×Bst DNA polymerase large fragment buffer solution (20 mMTris-HCl, 10 mM KCl, 10 mM (NH 4 ) 2 SO 4 , 2 mM MgSO 4 , 0.1% Triton X-100, pH 8.8) were mixed, and double-distilled water was added to make the total volume of the reaction system reach 50 μL. After the reaction was placed in a metal bath at 65 °C for 1 hour, the reaction was terminated at 95 °C for 10 minutes. The main difference from the existing hyperbranched rolling circle amplification reaction system is the use of the nicking endonuclease Nb.BsrDI. Mix 5 μL sample and loading buffer solution 6×loading buffer (1 μL) evenly, and pour into the sample well. Th...
Embodiment 3
[0037] Example 3: Atomic Force Microscopy Characterization of Amplified Products of Linear Rolling Circle Amplification, Hyperbranched Rolling Circle Amplification, and Network Rolling Circle Amplification
[0038] The amplification products of linear rolling circle amplification, hyperbranched rolling circle amplification and network rolling circle amplification were characterized by atomic force microscopy.
[0039] attached by Image 6 A It can be seen that the amplification product of the linear rolling circle amplification reaction is several linear single strands, which is different from the theoretical rolling circle replication reaction, that is, the extension product generated by hybridizing the target DNA to a circular single-stranded DNA molecule It is the multiple connections of the complementary pairing products of this circular DNA template molecule to form a certain length of linear single-stranded DNA.
[0040] attached by Image 6 B It can be seen that the a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com