Detection device for instability of genome copy number
An unstable and detection device technology, applied in the field of medical detection, can solve the problems of low specificity and accuracy, and it is difficult to determine the cancer discrimination standard.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Detection device for genome copy number instability based on high-throughput sequencing
[0030] A detection device for genome copy number instability, including a sample preparation unit, a sequencing unit, a calculation unit, and a statistical analysis unit, wherein:
[0031] The sample preparation unit includes disposable venous blood collection needle, disposable vacuum blood collection tube, blood anticoagulant EDTA, blood cell protection agent, plasma free DNA purification kit, which is used to extract peripheral blood from the vein of the subject and separate it into the plasma Cell-free DNA;
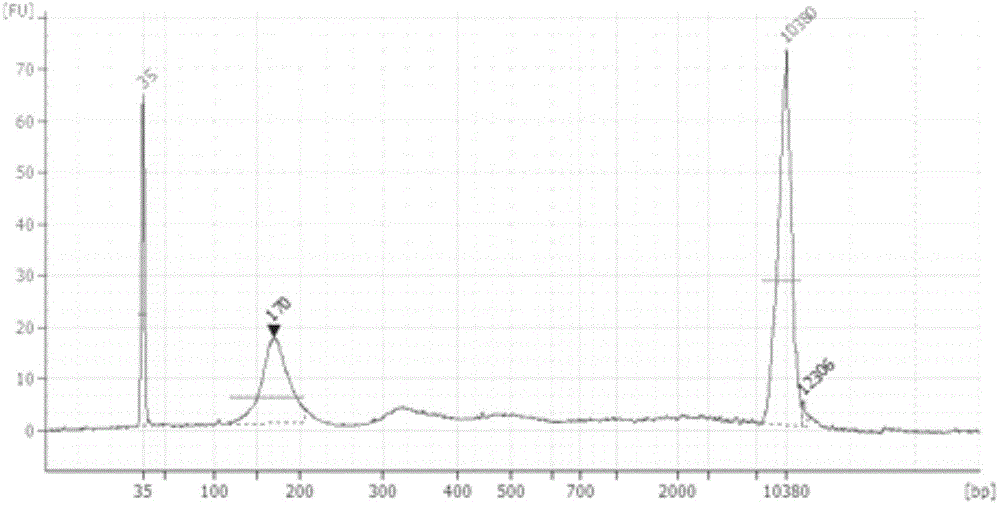
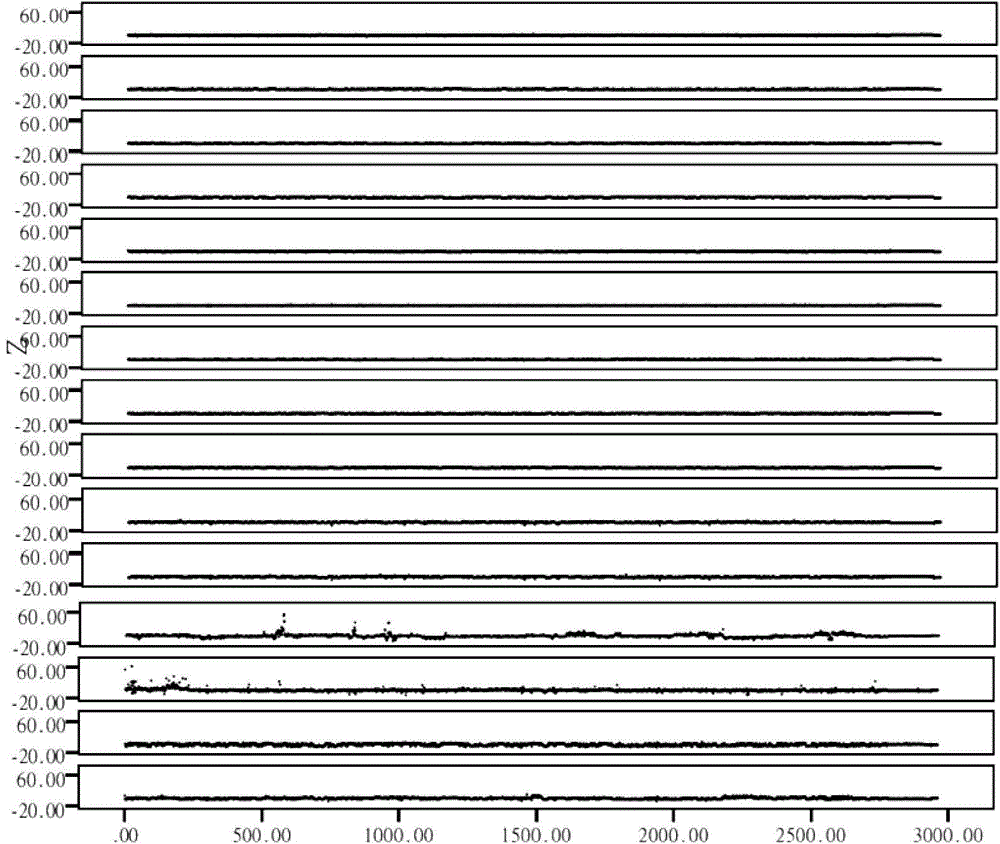
[0032] The sequencing unit includes a quality inspection module, a high-throughput sequencing module, and a data optimization module, and the high-throughput sequencing module includes a library construction kit and a high-throughput sequencing kit. The quality inspection module is used to perform quality inspection of DNA concentration and fragment size before ...
Embodiment 2
[0035] Example 2 Detection device for genome copy number instability based on gene chip
[0036] The detection device for genome copy number instability involved in this example has basically the same structural composition as the detection device for genome copy number instability in Example 1, and also includes a sample preparation unit, a sequencing unit, a calculation unit and a statistical The analysis unit is different in that: in the sequencing unit of the device of this embodiment, the high-throughput sequencing module in Example 1 is replaced by a gene chip module, and the gene chip module includes a gene chip and a fluorescent signal detector; The gene chip module performs hybridization and sequencing on the free DNA in the plasma sample to be tested, and detects the fluorescence signal intensity value of each genome window through a fluorescent signal detector, so as to quantify the number of sequencing bands in each genome window.
Embodiment 3
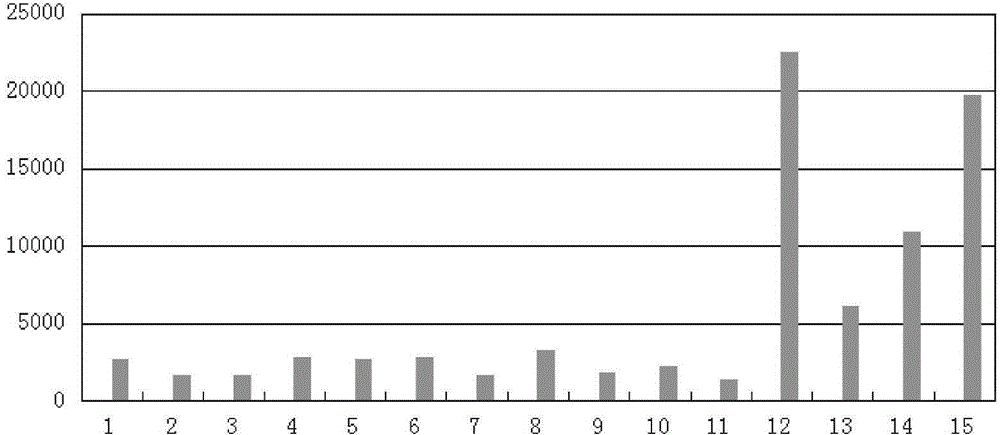
[0037] Embodiment 3 The test example of using the device of Embodiment 1 for cancer screening (statistical ZA value)
[0038] This example relates to a test example of using the high-throughput sequencing-based genome copy number instability detection device of Example 1 for cancer screening.
[0039] 1. Build the database for the database module
[0040] 1. Cut the human genome reference sequence GRCh38 into first-to-first-connected and non-overlapping genome windows, each genome window is 1Mb base pairs in size, and each genome window is an independent analysis unit.
[0041] 2. For the above-mentioned about 3,000 genome windows, according to their GC content, whether they contain genes, whether there are copy number variations, whether there are repeated sequences, how many single nucleotide polymorphism (SNP) sites, and whether there are known The characteristics of cancer-related genes and other characteristics are matched with weights respectively.
[0042] 3. Perform ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com