Detection method for CYP2C9*2 gene polymorphism, as well as nucleic acid probe and kit for method
A technology for gene polymorphism and detection probes, which is used in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0107] 1. Preparation of probes and primer pairs for CYP2C9*2 gene
[0108]Probes for CYP2C9*2 genes and primer pairs for CYP2C9*2 genes were prepared using conventional techniques.
[0109] The specific sequence is as follows:
[0110] Probe for CYP2C9*2 gene
[0111] *2 Mutant probe FAM-aacacAgtcctcaAtgctcc-BHQ2
[0112] *2 wild-type probe HEX-ttgaacacGgtcctcaatgctc-BHQ2
[0113] Primer pair for CYP2C9*2 gene
[0114] Forward primer sequence 3D1-S-1: AAGGCAGCGGGCTTCCTC
[0115] Reverse primer sequence 4D1-A-1: GCTGCGGAATTTTGGGATGG.
[0116] Second, configure the PCR reaction solution,
[0117] The serving size is as follows:
[0118] Commercially available 2×PCR reaction solution (including taq enzyme, UNG enzyme, dNTP, dUTP) 10 μL
[0119] Primer and probe mix 1 μL
[0120] Internal control probe mixture 1 μL
[0121] RNase-free water 7 μL
[0122]
[0123] Aliquot, 19 μl per tube into 0.2ml PCR reaction tubes / plates, and transfer to the sample processing area...
Embodiment 2
[0137] Steps one to four:
[0138] In the same method and steps as in Example 1, the DNA sample was replaced with a DNA sample expressing wild type CYP2C9*2, and 1 μl of the wild type DNA sample was added to 19 μl of the PCR reaction solution.
[0139] 5. Results analysis
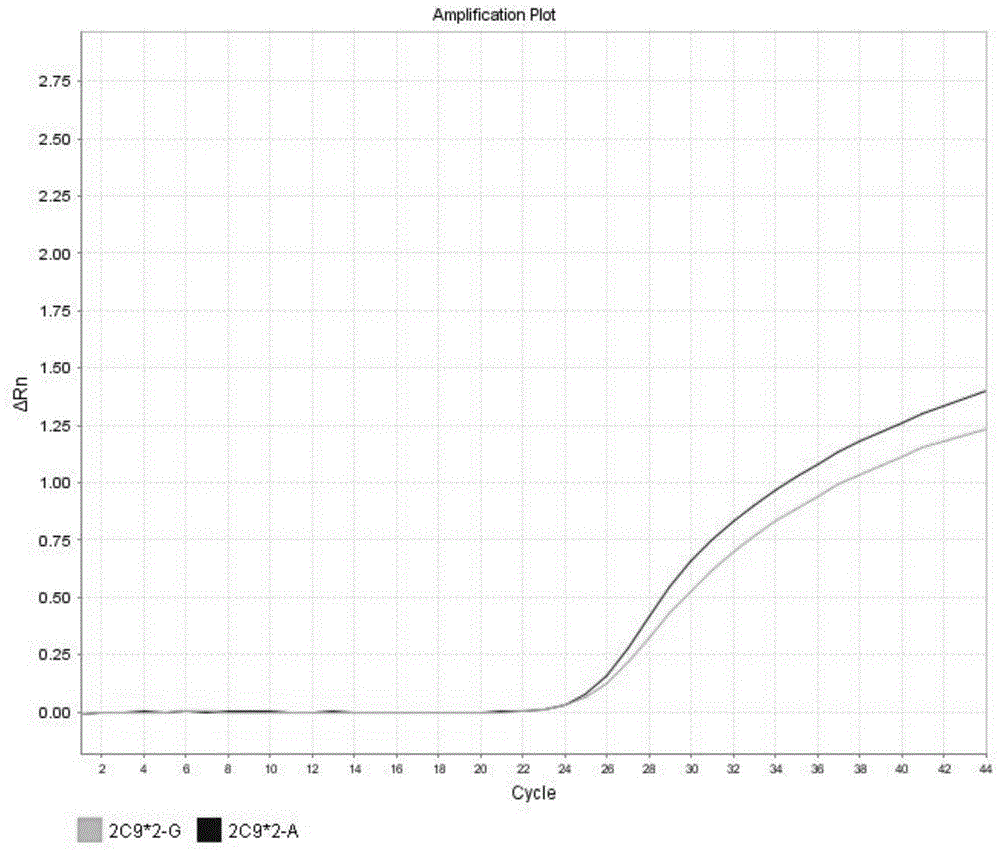
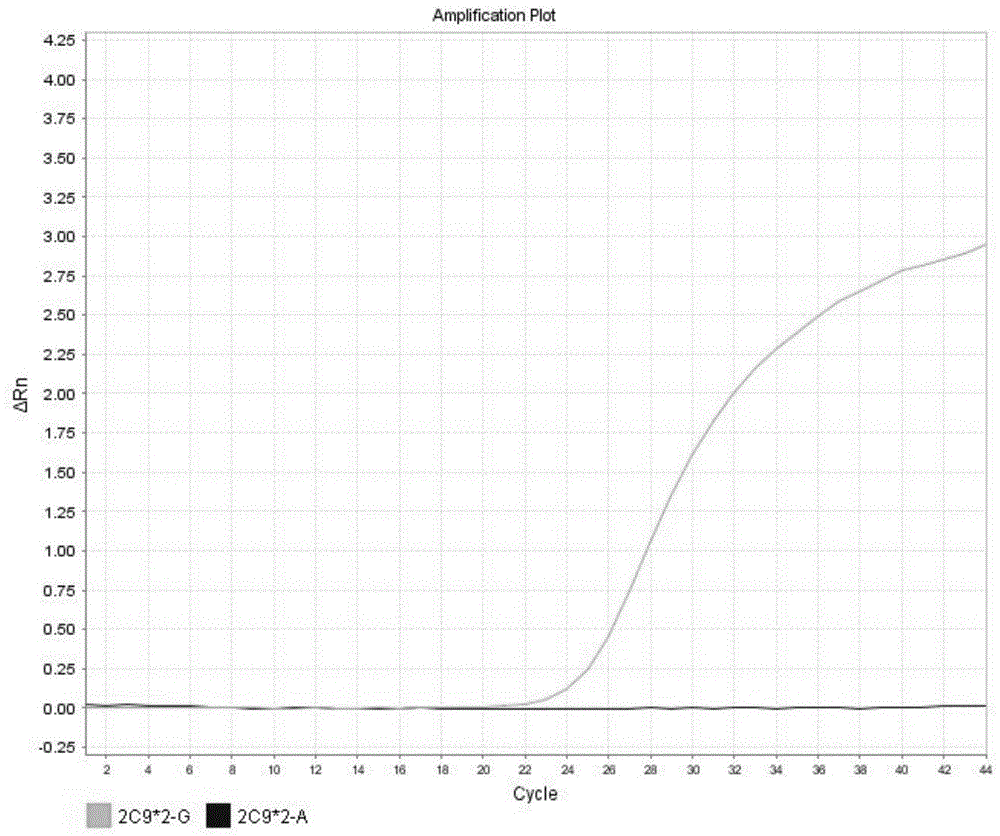
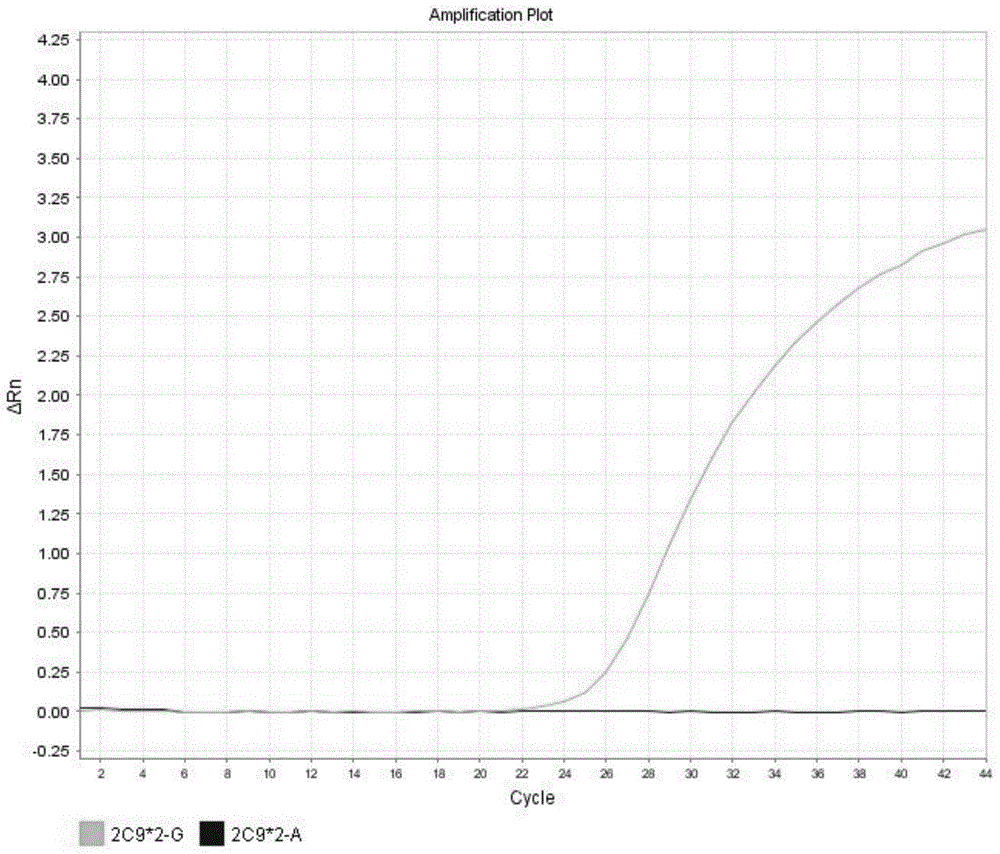
[0140] CYP2C9*2 expresses wild type results in figure 2 shown in the table. These graphs are analysis graphs showing changes in fluorescence intensity over time, that is, changes in fluorescence intensity as the number of amplification cycles increases. Such as figure 2 As shown, the fluorescence curve indicates that the CYP2C9*2 gene in the sample is wild type.
Embodiment 3
[0142] one,
[0143] 5 mL of venous blood was collected from 3 subjects A, B and C, and anticoagulated with EDTA (obtained samples 1, 2 and 3).
[0144] Methods of Using the RelaxGene Blood Genomic DNA Extraction Kit
[0145] 1. Add 3 mL of EDTA anticoagulated venous blood, add 5 mL of Lysis Solution CL to lyse blood cells, transfer the solution to a clean and sterile 15 mL centrifuge tube, invert and mix well; centrifuge at 2,000×g for 5 minutes at room temperature, discard the supernatant, and keep the precipitate ;
[0146] 2. Add 5ml lysate CL to the centrifuge tube, shake vigorously until the precipitate is completely dissolved, and centrifuge at 2,000×g for 5 minutes at room temperature;
[0147] 3. Discard the supernatant, add 2.5ml proteinase K digestion solution, vortex and mix; 56°C water bath for 10 minutes.
[0148] 4. Add 2.5ml of isopropanol, shake the centrifuge tube gently, and pick out the filamentous genomic DNA.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Pre-denatured | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com