Method for screening and detecting single-nucleotide polymorphic site G642A of marsupenaeus japonicus
A single nucleotide polymorphism, G642A technology, applied in the field of screening and detection of the G642A single nucleotide polymorphism site of Penaeus japonicus, can solve the problems of high cost and inability to directly apply the marker, and achieve simple and fast operation , The effect of low cost of screening and testing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] 1. Use bioinformatics analysis to obtain spliced sequences containing candidate single nucleotide polymorphic sites. The specific process is as follows: First, perform transcriptome sequencing of the hepatopancreas of the mixed sample Penaeus japonicus, and use trinity software to splice according to the default parameters Hepatopancreas reference transcripts, use BWA software to align transcriptome sequencing reads (reads) to reference transcripts according to the default parameters, use GATK software to screen for alignment quality scores greater than 40, minor allele frequency above 200, minor allele frequency If the locus with an allele frequency higher than 20% is a candidate locus, the comp11169 splice sequence is screened for primer design, and its sequence is:
[0026] GATGTCGTATTAATGTCTTTATCCAATTAAAAAATCATCATTTATAATTCTAAGCATCTGACGACTATAATATGTAGGTATAACATTGTACTTATACATGGAGAGTCGAAATCTTGAATCATTTAACCTGTTCCTACAATATCACAAGAGACAAGTAATTGAACCAATCACAATCACAAAAGTAAAGGAAAGAAA...
Embodiment 2
[0034] 1. According to the screening of comp11169 splicing sequence in Example 1, design sequencing primers for G642A_SNP site verification and typing.
[0035] 2. The PCR product amplified by the designed sequencing primers should include the single nucleotide polymorphism G642A_SNP site, and the primer sequence is:
[0036] Positive strand primer three: 5'-CGTGCCCTTGTAGATGCA-3';
[0037] Negative strand primer: 5'-TTCGTCGAGAGAACCACT-3'.
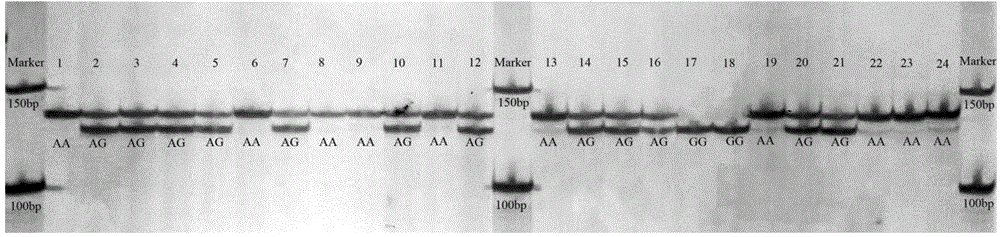
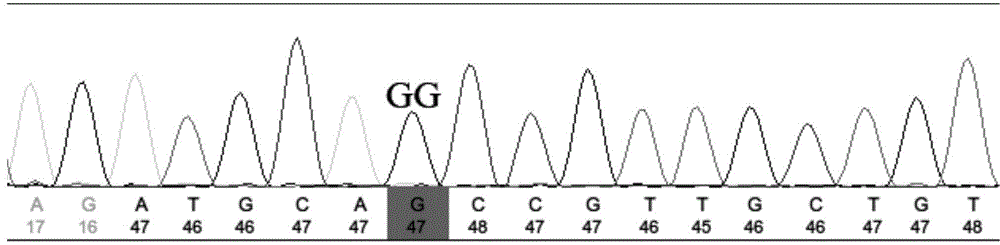
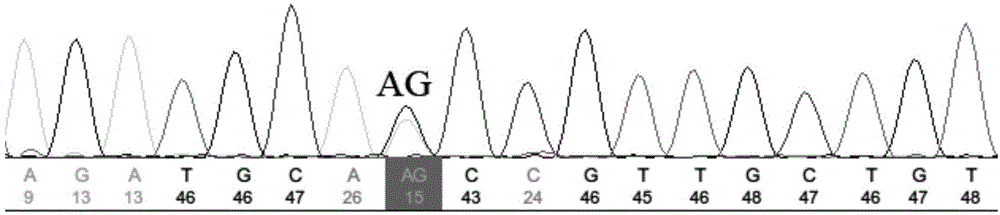
[0038] 3. Use the sequencing primers designed in step 2 to perform PCR amplification on the 24 individuals in Example 1. The PCR reaction system is 30 μL, including 1 μL of 50 ng / μL Genomic DNA Solution of Penaeus japonicus, 3 μL of 10×PCR buffer, 2.5 mM dNTP Mixes 2.4 μL, 25 mM MgCl 2 4 μL, 0.15 μL of 5U / μL Taq DNA polymerase, 0.5 μL of 10 uM forward-strand primer and 0.5 μL of reverse-strand primer, add ddH2O to 30 μL; PCR reaction program: denaturation at 94°C for 10 min; 94°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec , 40 cycles;...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com