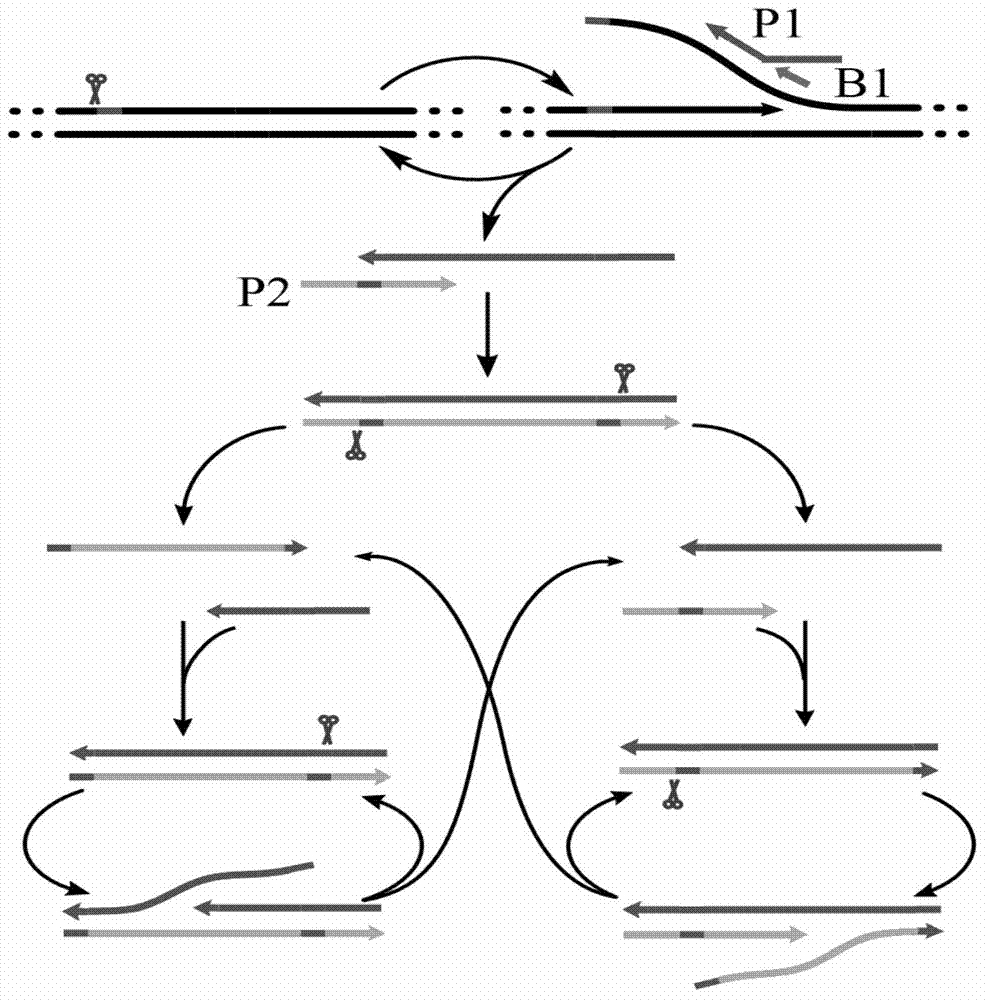
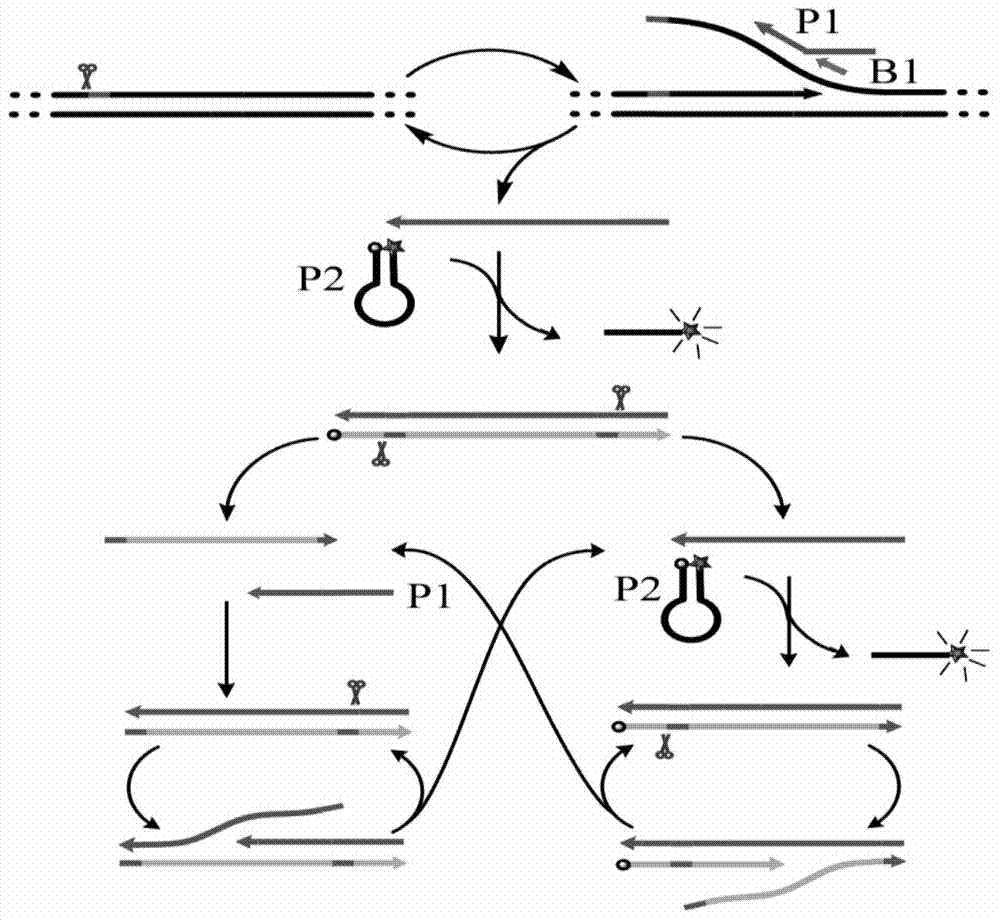
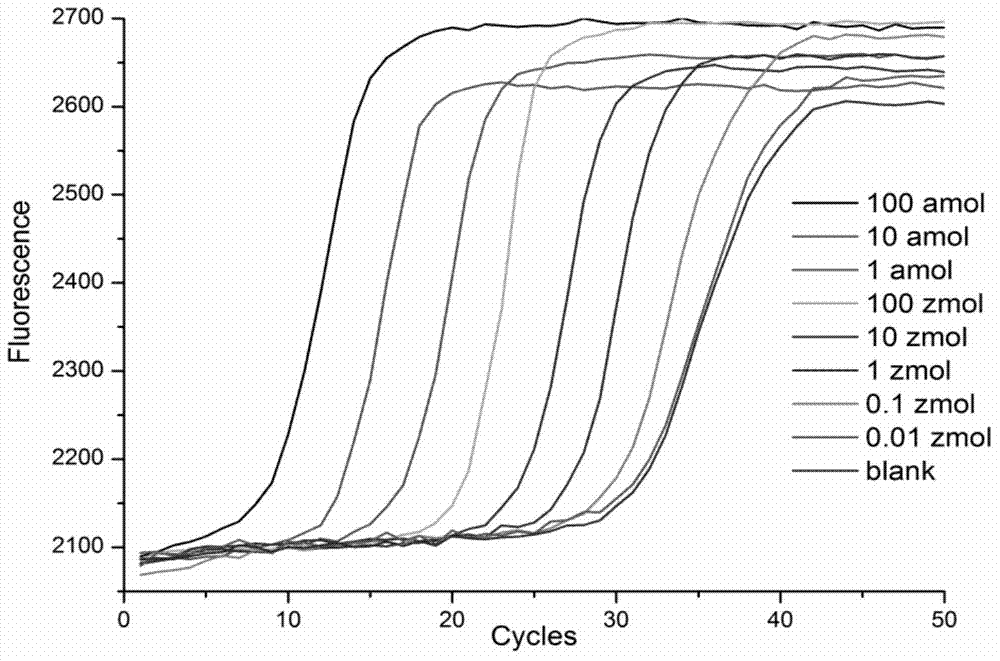
Novel method for isothermal amplification detection of double-stranded nucleic acid based on nicking enzyme
A double-stranded nucleic acid, nicking endonuclease technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of complex reaction system, non-specific amplification, replacement of primers to reduce amplification efficiency, etc. Achieve the effect of efficient amplification reaction, simplified operation, and short reaction time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Embodiment 1: Verify the feasibility of the method and the correctness of its principle. In this example, PBS plasmid is used as the target nucleic acid, three primers are used for constant temperature detection of exponential amplification, and the feasibility of the method and the correctness of the principle are verified by fluorescent signals. To the isothermal amplification system, 1 μL of target E. coli PBS plasmids of different concentrations (ie, SEQ ID NO.1 containing the amplified sequence of 5'-CATTGCTACAGGCATCGTGGTGTCACGCTCGTCGTTTGGTATGGCTTCATTCAGCTCCGGTTCCCAACGATC-3') and 1 μL of double distilled water were added as a negative control group, 10×Thermpol (200mM Tris-HCl, 100mM KCl, 100mM (NH 4 ) 2 SO 4 , 20 mM MgSO 4 , 1% Triton X-100pH8.8@25℃) 1μL, 0.1μL Bst DNA polymerase, large fragment, 0.5μL Nt.BsrDI nicking endonuclease, 0.6μL (2.5mM) dNTPs, 1μL (8×10 -6 M) PBS amplification primer PBS-P1 (that is, the sequence of SEQ ID NO.2 is 5'-CGCTACCCATACATAC...
Embodiment 2
[0065] Embodiment 2: the specificity of verification method
[0066] 100zmol of the target Escherichia coli PBS plasmid added to the isothermal amplification system (i.e., SEQ ID NO. 1 contains the amplified sequence of 5'-CATTGCTACAGGCATCGTGGTGTCACGCTCGTCGTTTGGTATGGCTTCATTCAGCTCCGGTTCCCAACGATC-3"), and then added different amounts of Escherichia coli genome (respectively 0zmol , 10zmol, 100zmol, 1amol), 10×Thermpol (200mM Tris-HCl, 100mM KCl, 100mM (NH 4 ) 2 SO 4 , 20 mM MgSO 4 , 1% Triton X-100pH8.8@25℃) 1μL, 0.1μL Bst DNA polymerase, large fragment, 0.5μL Nt.BsrDI nicking endonuclease, 0.6μL (2.5mM) dNTPs, 1μL (8×10 -6 M) PBS amplification primer PBS-P1 (that is, the sequence of SEQ ID NO.2 is 5'-CGCTACCCATACATACTGTTCCATTGGCCCATACCAAACGACGAG-3'), 1 μL (3×10 -7 M) PBS replacement primer PBS-B1 (that is, the sequence of SEQ ID NO.3 is 5'-GGAACCGGAGCTGAATG-3'), 1 μL (8×10 -6 M) PBS amplification primer PBS-P2 (that is, the sequence of SEQ ID NO.4 is 5'-CGCTACGGTTCTATAGTGT...
Embodiment 3
[0067] Embodiment 3: Using this method to detect Vibrio parahaemolyticus
[0068] After measuring the concentration of the extracted Vibrio parahaemolyticus genomic DNA, perform gradient dilution, and add different concentrations of Vibrio parahaemolyticus genomic DNA (that is, SEQ ID NO.5 contains the amplified sequence of 5 '-ACATTGCGTATTTTCGCAGGTCAAAAATGATCCAACAGACATCTGCAAATAAAGGAGGCCAGCATGAAGATTAAAGTAGCATCTGCGGTTTTGGCCGT ATCTA-3") 1 μL and as a negative control group, add 1 μL of double distilled water, 10×Thermpol (200mM Tris-HCl, 100mM KCl, 100mM (NH 4 ) 2 SO 4 , 20 mM MgSO 4 , 1% Triton X-100pH8.8@25℃) 1μL, 0.1μL Bst DNA polymerase, large fragment, 0.5μL Nt.BsrDI nicking endonuclease, 0.6μL (2.5mM) dNTPs, 1μL (8×10 -6 M) Vibrio parahaemolyticus amplification primer VP-P1 (that is, the sequence of SEQ ID NO.6 is 5'-ATCACGTCAGTCTACTCGTAGCATTGCCCTTTATTTGCAGATGTCTGTTG-3'), 1 μL (3×10 -7 M) Vibrio parahaemolyticus replacement primer VP-B1 (that is, the sequence of SEQ ID...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com