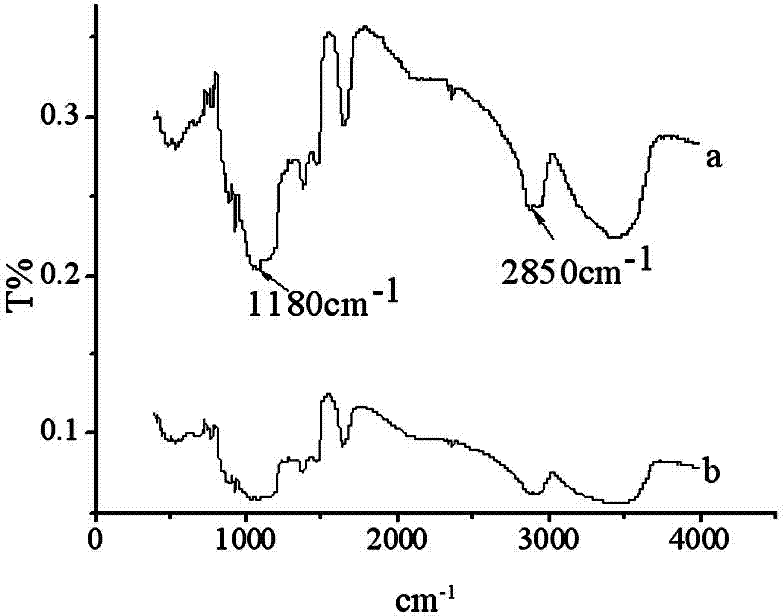
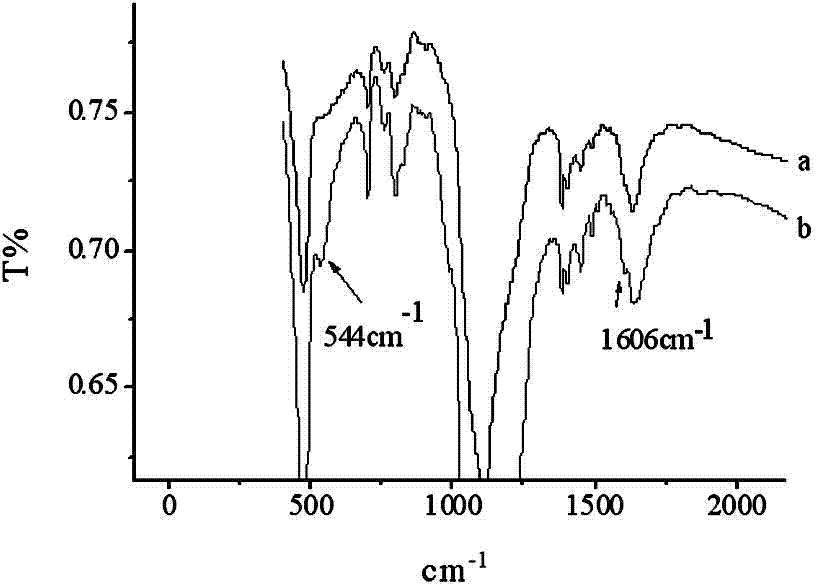
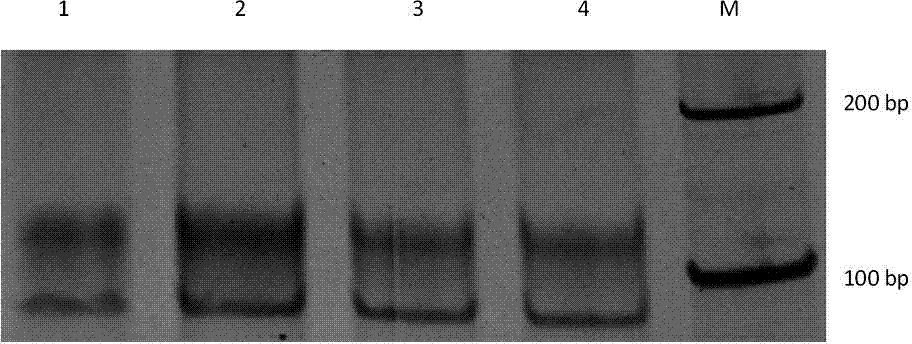
Specific nucleic acid aptamer for recognizing streptomycin and application of nucleic acid aptamer to streptomycin detection
A nucleic acid aptamer and specific recognition technology, which is applied in the field of biotechnology detection, can solve the problems of cumbersome detection process of streptomycin, poor accuracy and sensitivity, and cumbersome detection procedures, and achieve flexible methods, easy preservation, and low cost. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Acquisition of a nucleic acid aptamer that specifically recognizes streptomycin
[0037] (1) Coupling of epoxy-based gel and streptomycin: Take 3 ml of epoxy-based activated gel and wash it five times with double distilled water, then filter and dry; add the dried epoxy-based activated gel to 10 ml of 0.1mol / L Na containing 20 mg / ml streptomycin 2 CO 3 -NaHCO 3 In the coupling buffer (pH value 10.5), incubate at 37°C for 16 h; after the reaction, wash the incubated product three times with 0.1 mol / L PBS solution, then add 1 mol / L ethanolamine for blocking, and incubate at room temperature for 1 h ; The blocked gel conjugate was sequentially washed with 5 times deionized water, 0.1 mol / L HAc-NaAc buffer solution (pH 4.0) containing 0.5 mol / L NaCl, deionized water, 0.1 mol / L containing 0.5 mol / L NaCl boric acid-sodium tetraborate buffer solution (pH value 8.0) and deionized water were fully washed, dried by suction filtration, and unreacted ligands were remov...
Embodiment 2
[0066] Example 2 Qualitative and quantitative detection of streptomycin using aptamers
[0067] (1) Colloidal gold preparation: 20 mL of HAuCl with a mass ratio concentration of 0.01% 4 Heat to boil, and add 500 μL of sodium citrate with a mass ratio concentration of 1% while stirring. After the color changes, the solution continues to boil for 10 minutes, remove the heat source and stir until room temperature to obtain colloidal gold; the colloidal gold can be stored in a mass ratio concentration of 0.05% sodium azide, spare;
[0068] (2) Qualitative detection: add 100 nmol / L nucleic acid aptamer that specifically recognizes streptomycin to 50 μL colloidal gold, and incubate at room temperature for 1 h to prepare aptamer-labeled colloidal gold; the final concentrations are respectively Streptomycin at 25 nmol / L, 50 nmol / L, 100 nmol / L, 200 nmol / L, 400 nmol / L, 800 nmol / L, and 1600 nmol / L was added to aptamer-labeled colloidal gold at room temperature Incubate for 1 h; finally...
Embodiment 3
[0073] Quantitative detection of streptomycin residues in embodiment 3 milk and honey
[0074]Streptomycin-free honey and milk samples were purchased from local supermarkets, and streptomycin was quantitatively detected by standard recovery experiments. The specific operations were as follows:
[0075] Milk sample processing: Add 3 mL ethyl acetate and 3 mL water to 1 mL milk sample, vortex for 15 min; then centrifuge at 8000 rpm / min at 4°C for 15 min; the mixture is divided into three layers, and the aqueous layer is collected as the next One-step testing of samples;
[0076] Honey sample processing: honey is diluted five times with double distilled water, and used as a test sample;
[0077] A series of standard concentrations of streptomycin: 25nmol / L, 50nmol / L, 100nmol / L, 200nmol / L, 400nmol / L were added to the treated milk and honey samples respectively, and the quantitative detection method in Example 2 was used to detect the concentration of streptomycin in the samples. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com