Probe based on double signal amplification and application of probe
A dual signal amplification and probe technology, applied in the field of molecular bioinformatics, can solve the problems of high cost, limited detection conditions, and low selectivity of DNA molecular detection probes, achieve superior specificity, enhance signal, and improve The effect of sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
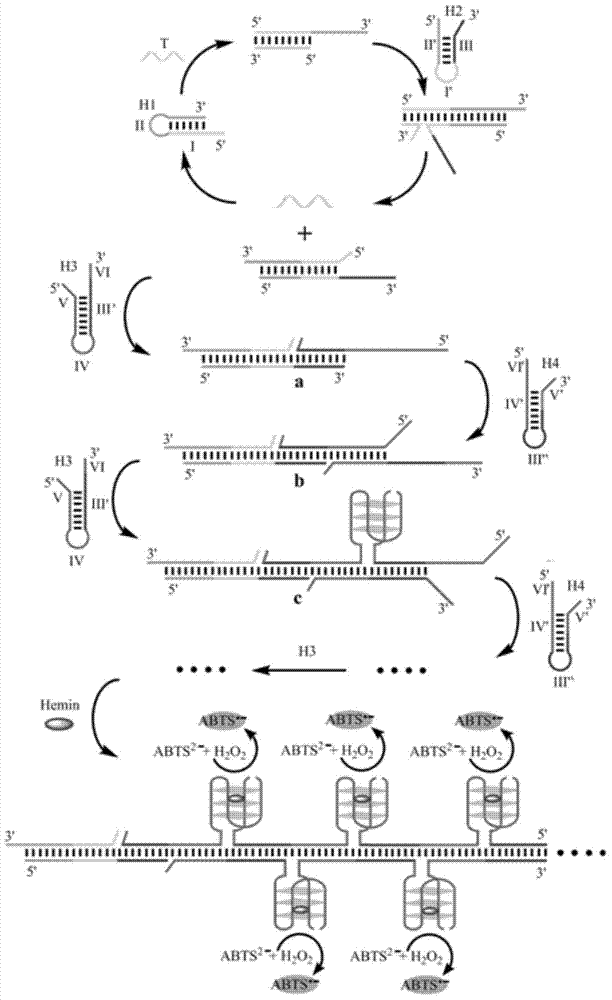
[0042] Embodiment 1: be used for detecting the design of the probe based on double signal amplification of HIV virus DNA molecule
[0043] In this embodiment, the probes based on dual signal amplification include: the first hairpin H1, the second hairpin H2, the third hairpin H3, and the fourth hairpin H4; the first hairpin H1, The second hairpin H2, the third hairpin H3, and the fourth hairpin H4 are all formed by the self-folding of the single-stranded linear molecule, and the complementary base pairing hybridization in the folded region, and the part of the double-stranded structure in the local region is the stem The stem region, the part that does not form a double-stranded structure and folds back is the loop region; the sequence of each component is shown in the table below. The target DNA molecule to be detected is HIV virus DNA molecule, which has the following sequence:
[0044] 5'-ACTGCTAGAGATTTTCCACAT-3'.
[0045] Table 1 Probe sequences based on dual signal ampl...
Embodiment 2
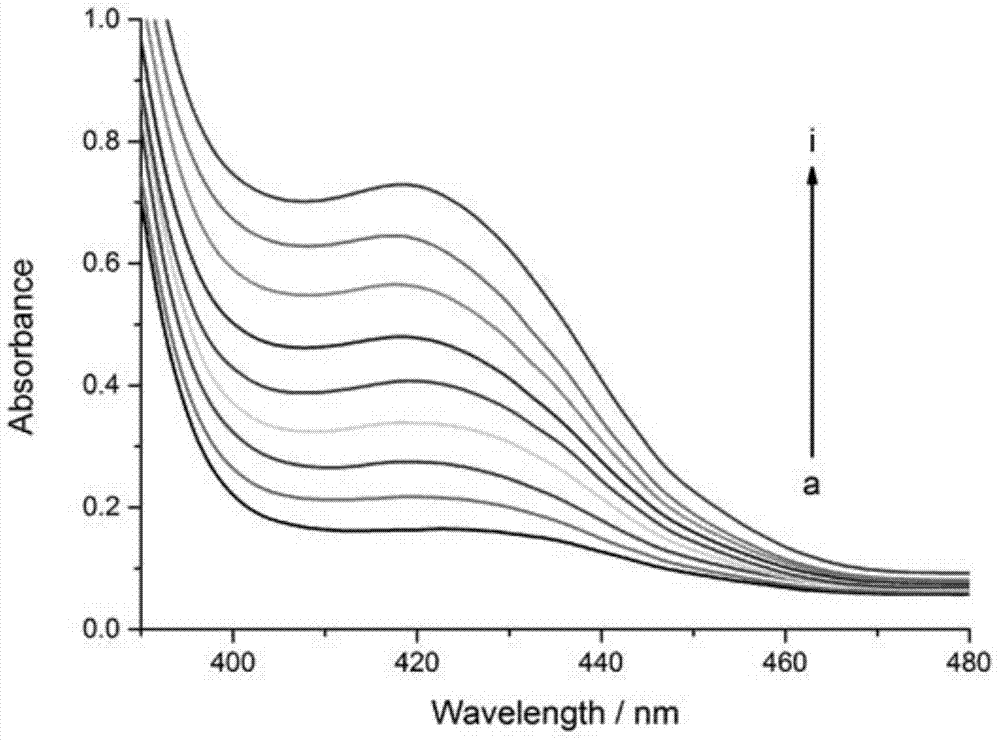
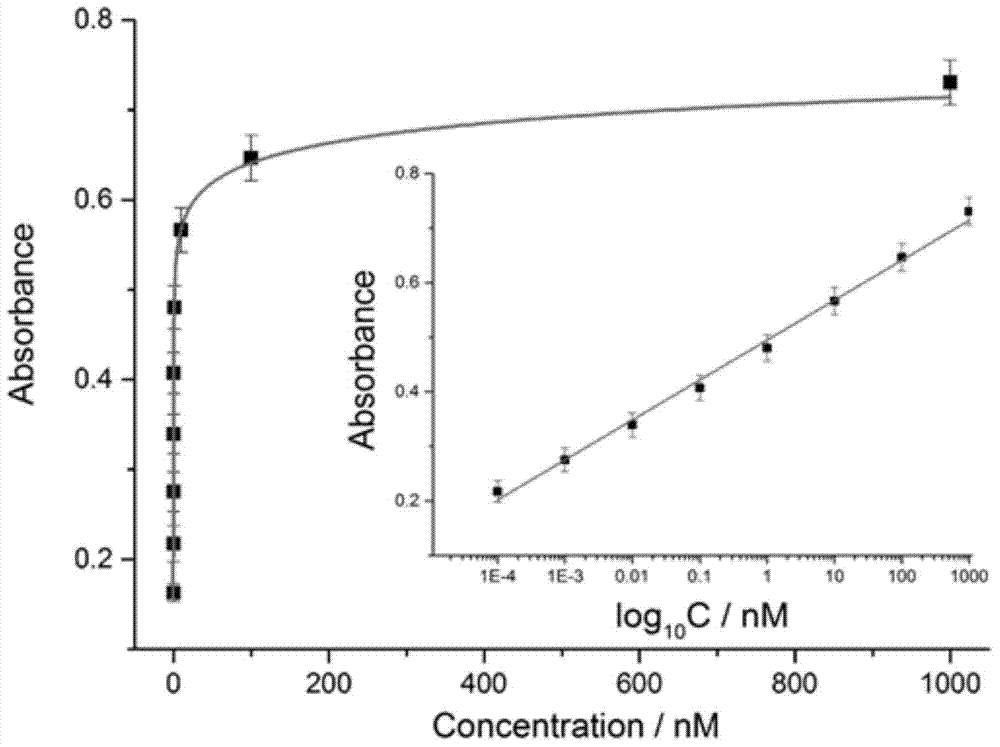
[0064] Example 2: Detection of target DNA molecules at different concentrations based on double signal amplification probes
[0065] The hemin used in this example, tris(hydroxymethyl)aminomethane (Tris), N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid (HEPES), 2,2'- Nitrogen-bis(3-ethylbenzothiazole-6-sulfonic acid) diammonium salt (ABTS) and hydrogen peroxide were purchased from Aladdin Reagent Company (Shanghai, China), and other chemical reagents were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China), there is no difference in the effect between the products of the various commercially available models of the above materials. All solutions were prepared using redistilled water prepared by a Milli-Q purification system (Billerica, Massachusetts, USA) with a resistivity of 18 MΩcm.
[0066] The detection of the target DNA in embodiment 1 comprises the following steps:
[0067] First, take the solutions of the first hairpin, the second hairpin, the t...
Embodiment 3
[0073] Example 3: Signal Amplification Verification Experiment of Probes Based on Double Signal Amplification
[0074] In this example, two DNA sequences G1 and G2 are designed to replace the third hairpin H3 and the fourth hairpin H4 in Example 1.
[0075] The sequence of the DNA sequence G1 is:
[0076] The sequence of the DNA sequence G2 is:
[0077] S1 is a 3 / 4 G-quadruplex sequence, and S2 is a 1 / 4 G-quadruplex sequence.
[0078] Its detection principle is as Figure 4 Shown as:
[0079] After the target DNA is recognized, the first hairpin H1 is opened, and the opened first hairpin H1 and the second hairpin H2 are hybridized, so that the target DNA is separated from the first hairpin by the second hairpin H2 through the process of DNA branch migration. Replaced on H1. The replaced target DNA can participate in the next round of hairpin self-assembly process. At the same time, the newly formed H1-H2 duplex narrows the distance between the split G-quadruplex frag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com