Fatty-liver-related liver cancer model building method based on knockout mice
A technology of knockout mice and construction methods, applied in the fields of life science and biology, can solve the problems of complex modeling, inability to replicate human models, and long modeling time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
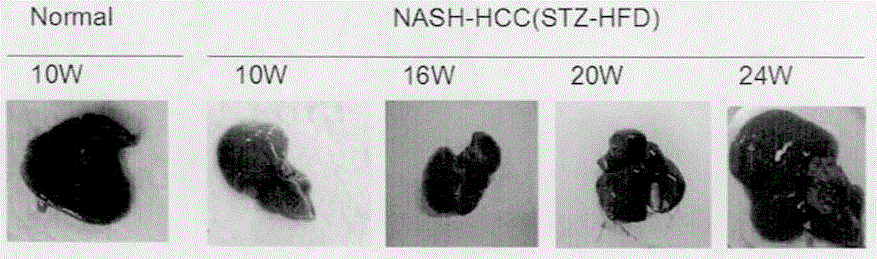
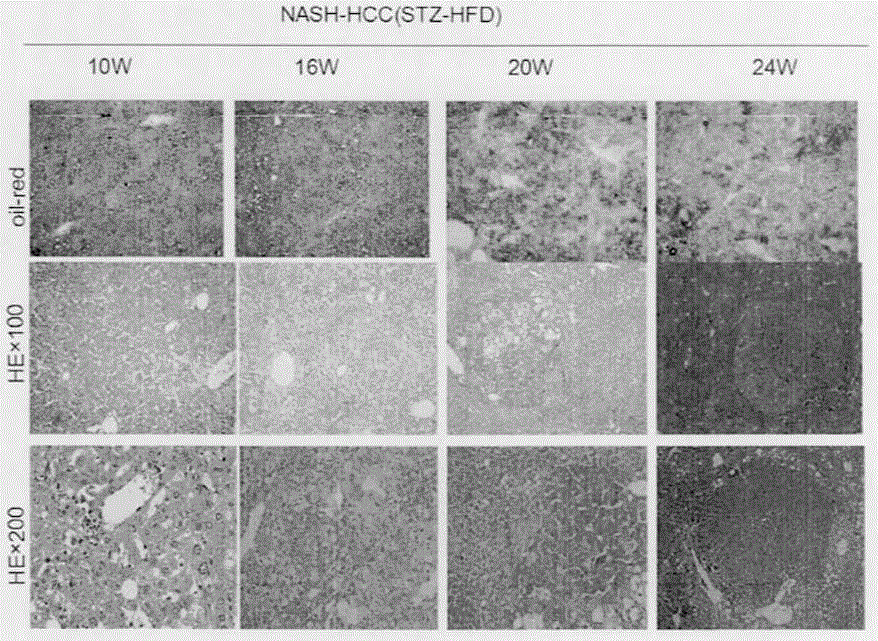
[0026]A method for constructing a fatty liver-related liver cancer model based on gene knockout mice. The main design idea is as follows: select APOE- / - and LDLR- / - gene knockout mice, including female mice and male mice, / - Gene knockout mice were purchased from the Institute of Model Animals, Nanjing University, Cat. No.: 002052, 002207, C57BL / 6J strain, the female and male mice were rationed to obtain double gene knockout mice, and the tail of the 1 mm mouse was cut off. Add 250 μl lysate for digestion, extract mouse genomic DNA, design primers: ApoE common forward primer: 5'-GCCTAGCCGAGGGAGAGCCG-3'; wild-type reverse primer: 5'-TGTGACTTGGGAGCTCTGCAGC-3'; deletion mutant reverse Primers: 5'-GCCGCCCCGACTGCATCT-3'.LDLR common forward primer 5'-CCATATGCATCCCCAGTCTT-3'; deletion mutant reverse primer: 5'-AATCCATCTTGTTCAATGGCCGATC-3'; wild type reverse primer: 5'-GCGATGGATACACTCACTGC-3 ', identify mouse genotype by polymerase chain reaction, the PCR reaction system is 12.5 μl of...
Embodiment 2
[0027] Embodiment 2 (control experiment) is based on APOE- / -gene knockout mouse NASH model
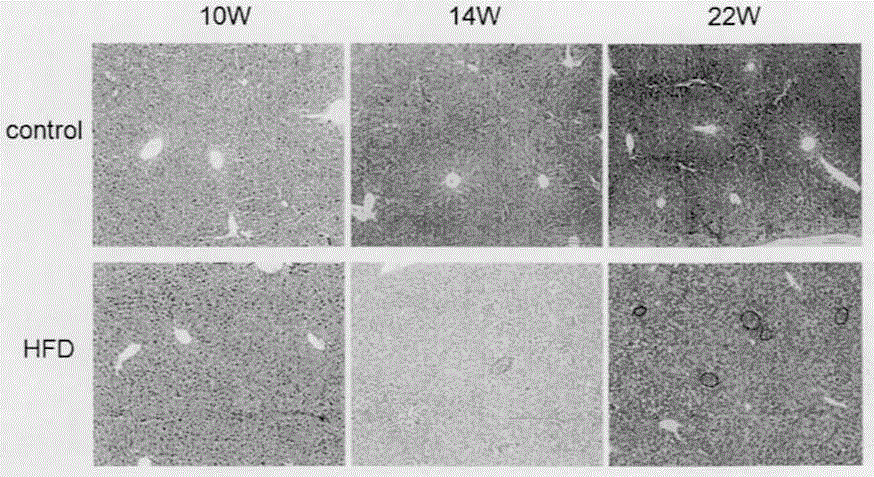
[0028] This experiment shows the HE staining results of 6-week-old APOE- / - knockout mice fed with high fat for 4 weeks, 8 weeks, and 16 weeks ( image 3 ). from image 3 It can be seen that the gene knockout mice only showed NASH at the 22nd week without STZ injection, and it took a long time to develop liver cancer. It has been pointed out in the literature that APOE- / - and LDLR- / - double gene knockout mice can spontaneously develop liver cancer after 35 weeks of high-fat diet. Our model can not only simulate the process of human NASH directly developing into HCC without fibrosis, but also has the advantages of short cycle, high induction rate, and convenient drug intervention and efficacy analysis.
[0029] Beneficial effects of the present invention: the present invention has been identified in two high-fat diets after weaning, that is, fatty liver begins to appear at 6 weeks, NA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com