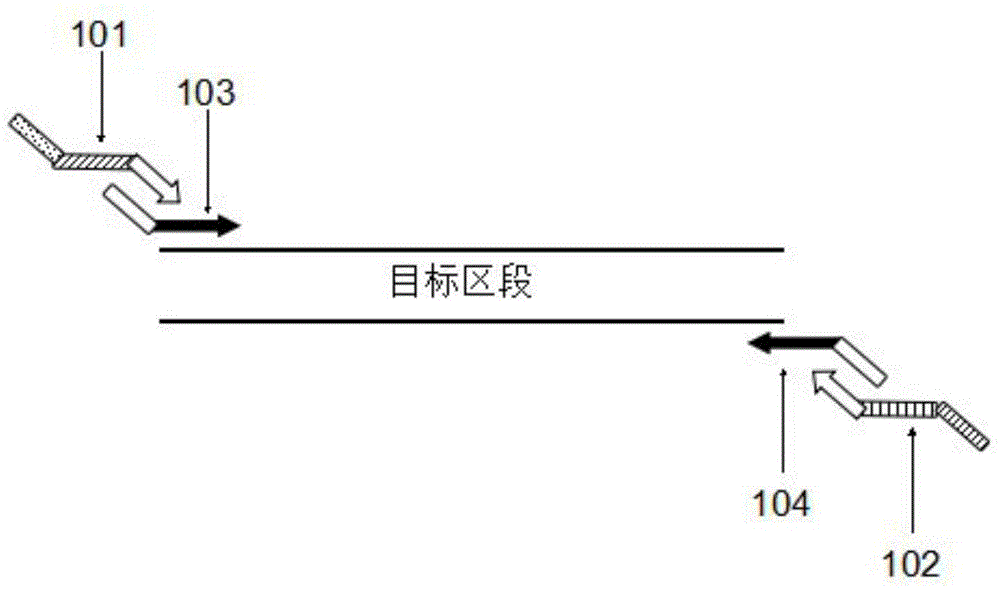
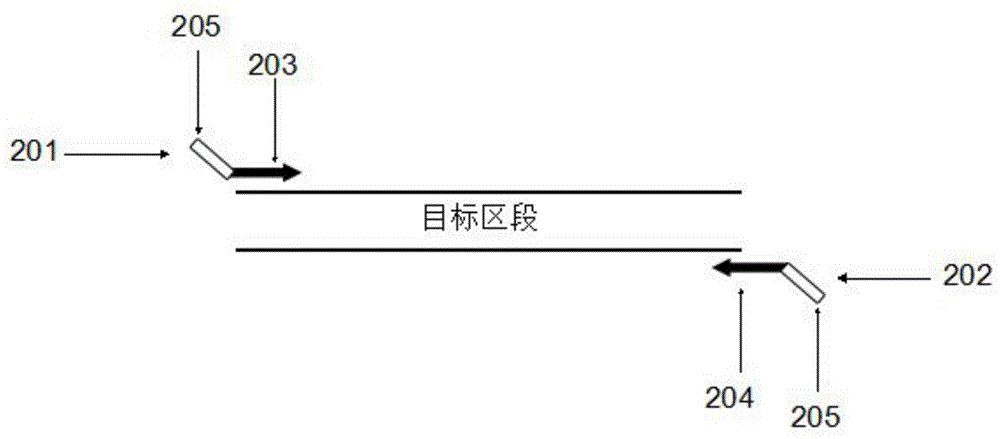
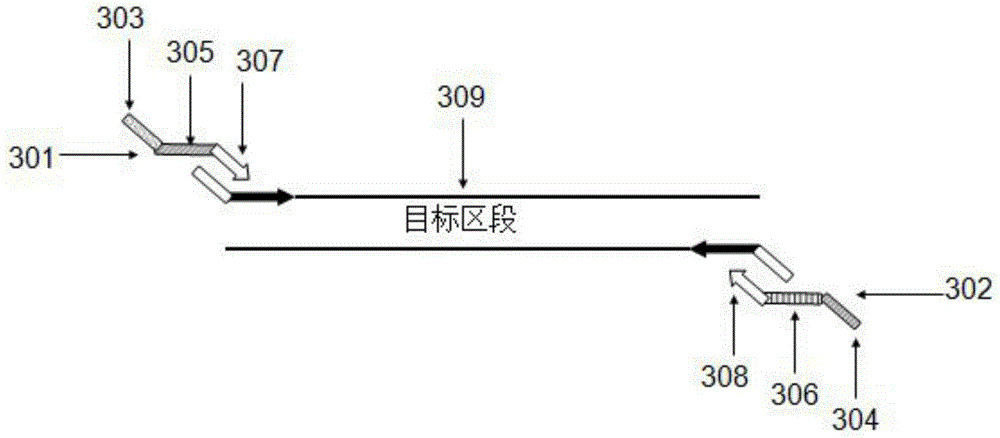
Multiplex-PCR three-round amplification method
A three-round, multiplex technology, applied in biochemical equipment and methods, recombinant DNA technology, and microbial assay/inspection, etc., can solve the problems of complex operation process, poor specificity, and unevenness of different samples and sites. Achieve increased efficiency and resolution, save time, and mitigate uniformity issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] IontorrentPGM platform was used to detect single nucleotide polymorphism (singlenucleotide polymorphism, SNP).
[0044] Sequence search and primer design: According to the SNP database of NCBI, a total of 37 target segment sequences carrying SNP sites in the human genome were found, and Primer3 online software was used to design primers according to general rules, and the upstream and downstream of the designed primers were artificially 5 The upstream universal sequence and the downstream universal sequence were added respectively at the 'end, and then the upstream and downstream specific primers were synthesized by chemical methods (Table 1).
[0045] Table 1. Specific primers for SNP detection
[0046]
[0047]
[0048]
[0049] DNA extraction: 757 groups of human blood cells were extracted using the blood genome miniprep kit according to the instructions to obtain DNA, and the DNA quality and concentration were determined by 0.8% agarose gel electrophoresis....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com