Space transcriptome database building and sequencing method and device adopted for same
A technology of spatial transcription and spatial location information, which is applied in the fields of medicine, biology, and agronomy, and can solve problems such as contamination of sequencing quality, sample loss, and inaccurate single-cell sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
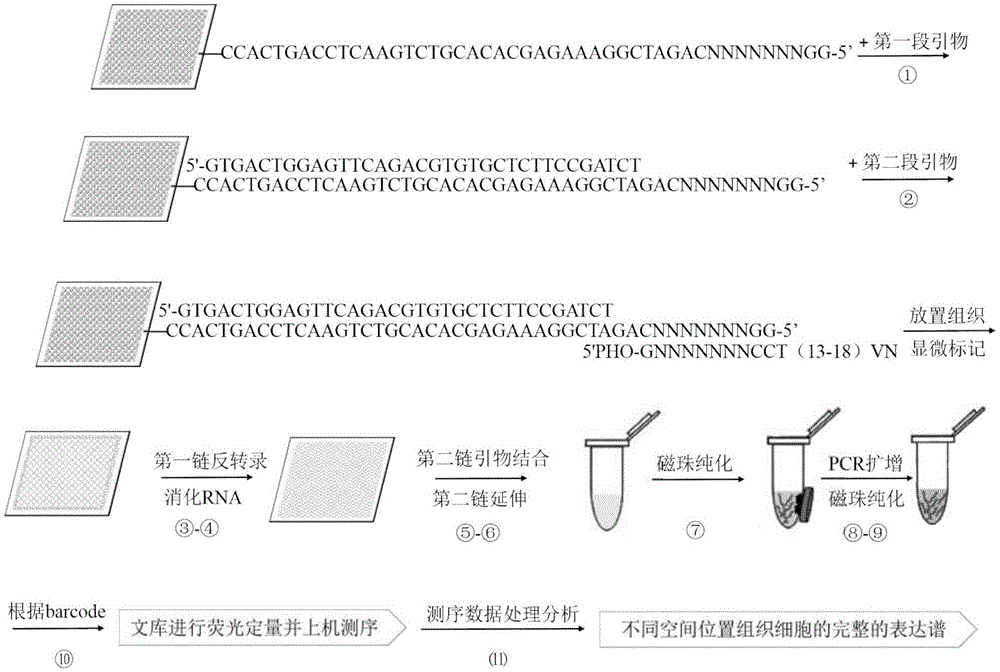
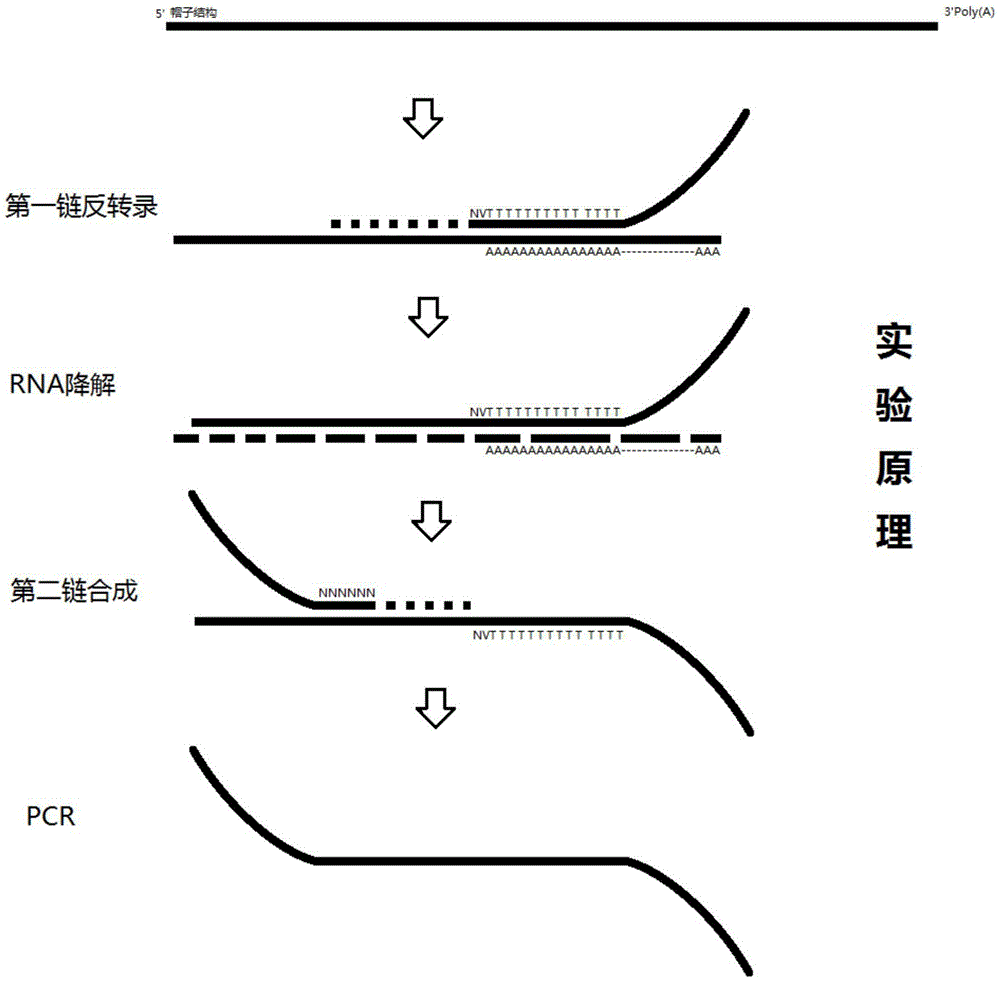
[0105] 1. Chip design and customization
[0106] Using the OligoArray in situ synthesis method, you can also use the in situ synthesis methods of other companies such as Affimatrix. The Oligoarray chip selects a 12k density synthetic chip, selects a characteristic sequence of 7 bases as a barcode marker, and removes all sequences containing 4 consecutive identical bases during barcode selection. Theoretically, the barcodes of adjacent sites differ by at least 4 bases, and the total amount of probes on the chip after synthesis is about 200ng.
[0107] The probe sequence synthesized by the chip is: the chip anchor probe is 5'-GGNNNNNNNCAGATCGGAAGAGCACACGTCTGAACTCCAGTCACC.
[0108] The sequence: AGATCGGAAGAGCACACGTCTGAACTCCAGTCACC is the reverse complementary sequence of the general adapter sequence P7 of Illumina Company.
[0109] The 7 Ns in GGNNNNNNNC represent 12,000 barcode sequences used to distinguish site information; due to the in-situ synthesis method, its 3' end is c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com