Nucleotide sequence, specific primer and method for identifying physalis angulata
A nucleotide sequence and specific technology, applied in biochemical equipment and methods, microbiological measurement/inspection, DNA/RNA fragments, etc., can solve the problems of safe use and protection of bitter medicinal resources, and the method is simple , short time effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1: Preparation of the characteristic nucleotide sequence of Solanum oleifera
[0023] 1. Genomic DNA Extraction
[0024] Cut Physalis plant samples (see Table 1) and put 100mg of fresh leaves into a mortar, immediately add liquid nitrogen to grind to powder, and then use UNIQ-10 column plant genomic DNA extraction reagent from Shanghai Sangon Bioengineering Co., Ltd. Genomic DNA was extracted from the box, and the resulting DNA was detected by electrophoresis with 0.8% agarose gel, and the DNA concentration was detected with an ultraviolet spectrophotometer, and diluted to 50 ng / μl.
[0025] Table 1. Physalis samples used in invention experiments
[0026]
[0027]
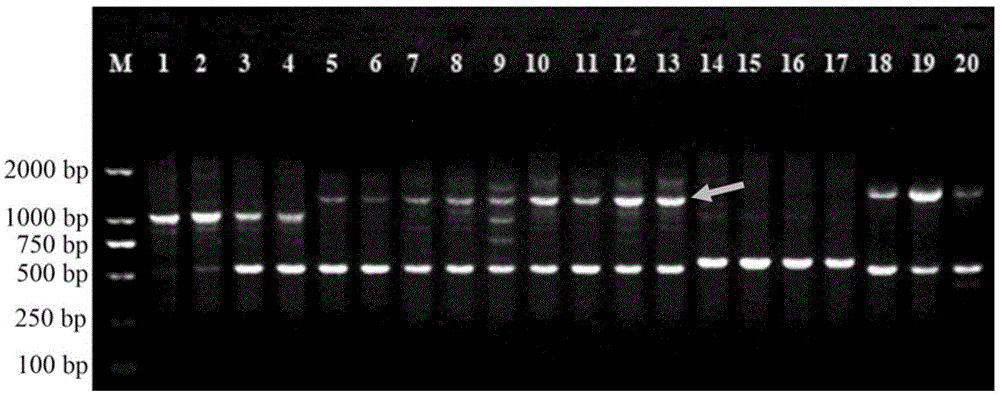
[0028] 2. SCoT–PCR reaction, electrophoresis detection
[0029] Use SCoT universal primer 3 (5'-CAACAATGGCTACCACCG-3') for PCR amplification, the primer was synthesized by Shanghai Sangon Bioengineering Co., Ltd., the reaction system (total volume 20μl) is: 10×Buffer2μl, MgCl2 (25mM) 2μl, dNT...
Embodiment 2
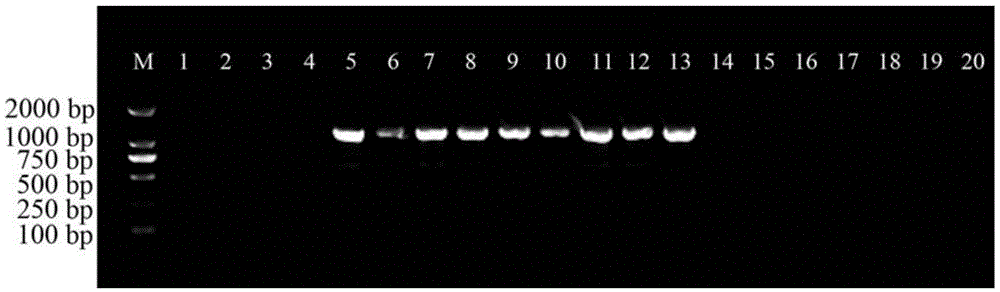
[0034] Example 2: Preparation of the specific primer ST3KZF / ST3KZR, PCR amplification, electrophoresis detection
[0035] On the basis of obtaining the specific nucleotide sequence of Sophora japonica, the nucleotide sequence of ST3KZF / ST3KZR (respectively shown in SEQIDNO.2 and SEQIDNO.3) was designed using PrimerPrimer5.0 software, and the primers were provided by Shanghai Sangong Synthesized by Bioengineering Ltd. Then, the designed and synthesized primers ST3KZF / ST3KZR are used to amplify and detect different Physalis plant samples (see the accompanying drawings for details).
[0036] The PCR reaction system (total volume 20μl) is: 10×Buffer2μl, MgCl 2 (25mM) 2μl, dNTPs (10mM) 0.8μl, primer ST3KZF (10μM) 1μl, primer ST3KZR (10μM) 1μl, template DNA (50ng / μl) 1μl, Taq enzyme (2U / μl) 0.5μl, ddH 2 O 11.7 μl.
[0037] The PCR reaction program was pre-denaturation at 94°C for 5 min; 35 cycles (denaturation at 94°C for 50 s, annealing at 65°C for 50 s, extension at 72°C for 1....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com