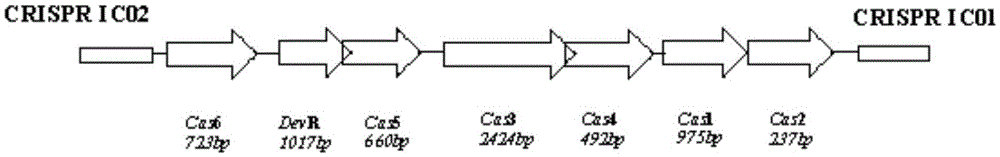
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat sequences)-Cas (CRISPR-associated proteins) system in Streptomyces virginiae IBL14 and method for carrying out gene editing by using CRISPR-Cas system
A technology of IBL14, gene editing, applied in the field of gene editing, to achieve the effect of improving production level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1 (svu016 gene knockout)
[0048] (1) Gene svu016 primer design and DNA amplification
[0049] According to the whole genome sequencing information, gene svu016 specific primers 016-F and 016-R were designed (Table 3). Genomic DNA of Streptomyces virginia IBL-14 was extracted, and PCR amplification of svu016 gene was performed using PfuDNAPolymerase produced by Shanghai Sangon Bioengineering Co., Ltd., reaction conditions: 95°C for 5min, 94°C for 30s, 58°C for 30s, 72°C for 2min , 2.5U PfuDNAPolymerase (50μl reaction system) produced by Sangon, 30 cycles, 72°C for 10min. The PCR product was detected by 1% agarose electrophoresis, the kit was recovered, and the purified svu016 full-length gene fragment was obtained for future use.
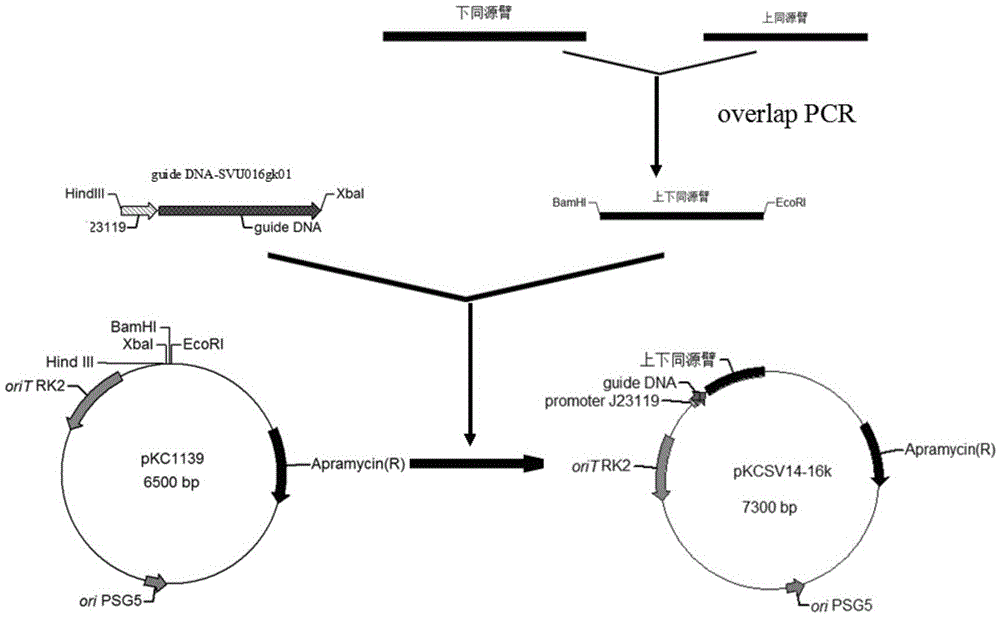
[0050] (2) Preparation of upstream and downstream homology arms
[0051] According to the full sequence of the svu016 gene (Table 2), the upstream homology arm primers 016-UF and 016-UR of the svu016 gene, and the downstream hom...
Embodiment 2
[0065] Embodiment 2 (insertion of chloramphenicol resistance gene)
[0066] (1) Gene svu016 primer design and DNA amplification
[0067] With embodiment 1 step (3).
[0068] (2) Preparation of upper and lower homology arms and chloramphenicol gene
[0069] The amplification of the upstream and downstream homology arms is the same as step (2) of Example 1. Chloramphenicol gene primers cm-F and cm-R (Table 3). Use the purified chloramphenicol gene DNA as a template to amplify the chloramphenicol gene. The reaction conditions are: 95°C for 5min, 94°C for 30s, 60°C for 30s, 72°C for 1min, 2.5U PfuDNA Polymerase (50μl reaction system ), 30 cycles, 10min at 72°C. The PCR product was detected by 1% agarose electrophoresis, recovered by the kit, and the purified chloramphenicol DNA fragment was obtained for future use.
[0070] (3) Preparation of editing template fragments
[0071] Mix 0.4 μl of the purified product of the upper homology arm and the purified product of the chlor...
Embodiment 3
[0082] Embodiment 3 (svu016 gene traceless point mutation)
[0083] (1) Gene svu016 primer design and DNA amplification
[0084] With embodiment 1 step (3).
[0085] (2) Preparation of point mutation editing template fragments
[0086] The point mutation editing template fragment SVU016C352T was directly synthesized by Chuzhou General Biology Co., Ltd., with BamHI and EcoRI restriction sites added at the beginning and end respectively. On this editing template fragment, the cysteine codon sequence at position 352 of the protein active site was mutated into Su amino acid codon sequence, and in order to ensure correct targeting, the amino acid codon sequence near the active site was modified to its degenerate sequence (Table 3).
[0087] (3) Preparation of targeted gene fragments
[0088] The target gene guideDNA-SVU016C352T was prepared in the same way as in step (4) of Example 1. (table 3)
[0089] (4) Construction of gene editing plasmid pKCSV14-SVU016C352T
[0090] W...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com