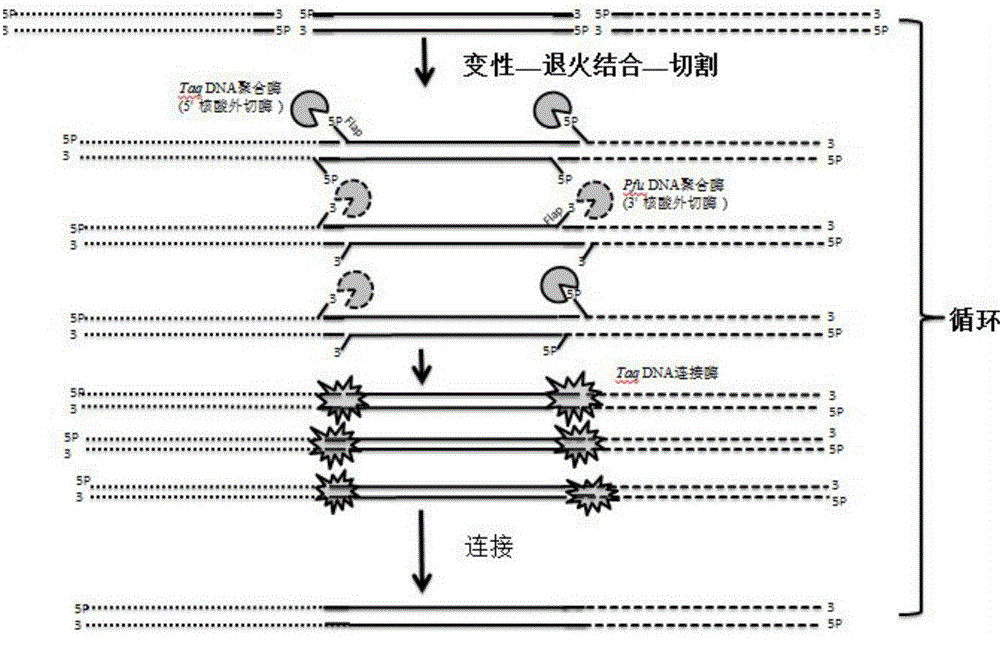
Method for rapidly and seamlessly assembling DNA in vitro on basis of heat-proof DNA polymerase and ligase
A technology of DNA ligase and polymerase, applied in the field of genetic engineering, can solve the problems of increased cost, complexity, and time-consuming of primer synthesis, and achieve the effect of rapid directed evolution and synthetic biology operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1 is optimized to the reaction system of DATEL assembly technology
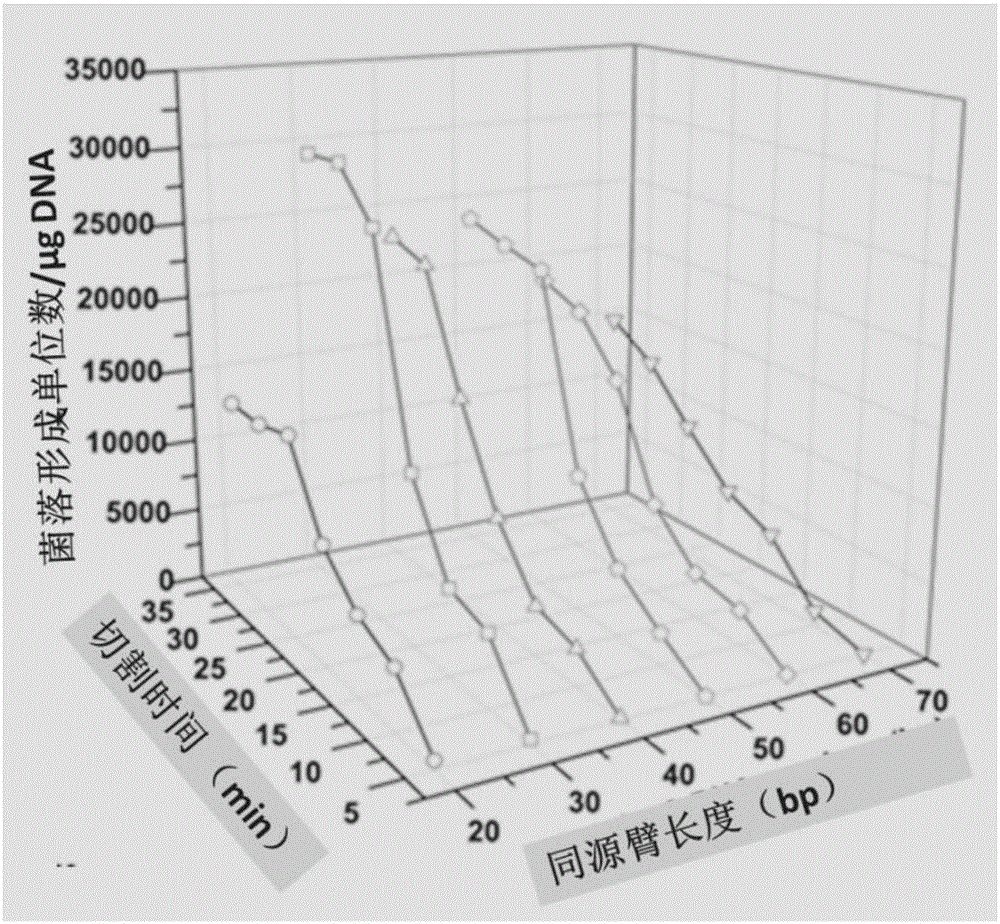
[0044] Taking the green fluorescent protein gene gfp and pUC19 vectors as examples, the assembly reaction was optimized. First, optimize the length of the "homologous arm" of the assembly reaction and the cutting reaction time, and design 6 sets of gfp amplification primers with 20bp, 30bp, 40bp, 50bp, 60bp and 70bp homology arms according to the gfp fragment and the carrier sequence, respectively, And a pair of vector pUC19 amplification primers. Primers are as follows:
[0045] TEL / puc19-F: TTCTTCTCCCTTACCCATGGCGTAATCATGGTCATAGCTGTTTCCT
[0046] TEL / puc19-R: TGGATGAACTATACAAATAACTGGCCGTCGTTTTACAACGTCG
[0047] TEL / gfp-P20F: CCATGGGTAAGGGAGAAGAACTTTTCAC
[0048] TEL / gfp-P20R: TTATTTGTATAGTTCATCCATGCC
[0049] TEL / gfp-P30F: CATGATTACGCCATGGGTAAGGGAGAAGAA
[0050] TEL / gfp-P30R: CGACGGCCAGTTATTTGTATAGTTCATCCA
[0051] TEL / gfp-P40F: CAGCTATGACCATGATTACGCCATGGGTAAGGGAGAAGAA
[0052] TEL / ...
Embodiment 2
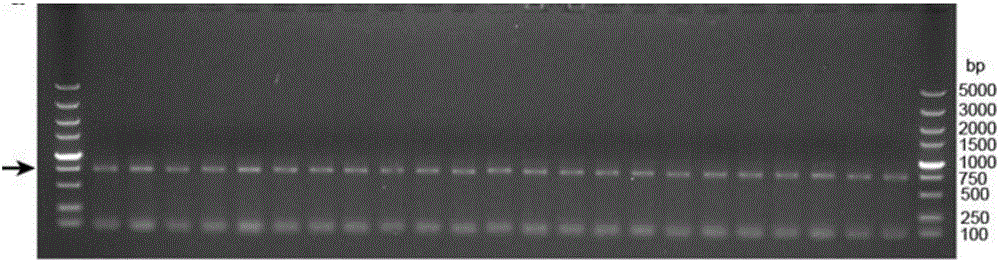
[0063] Embodiment 2 uses DATEL to quickly assemble multiple fragments
[0064] In order to verify the efficiency and accuracy of DATEL assembly of multiple fragments, the constitutive promoter PrpoS of the rpoS gene of Escherichia coli, the esterase gene estC23, the green fluorescent protein gene gfp, the kanamycin gene kan and the pbluesscriptIISK(+) vector were taken as examples , for assembly of 3-5 fragments ( Figure 4 a). All primers were first phosphorylated. The primer information is as follows:
[0065] MFA / pSK-R: GGTAATGGCAGTCGTGACTGGGAAAACCCTGGCGTTAC
[0066] MFA / pSK-4F:CTCGATGAGTTCTTCTAACCTGTGTGAAATTGTTATCCGCTCAC
[0067] MFA / pSK-3F: GTTCATCCGCGCCAACGCCGAGTAACCTGTGTGAAATTGTTATCCGCTCAC
[0068] MFA / pSK-2F:ATTACACATGGCATGGACGAACTATACAAATAACCTGTGTGAAATTGTTATCCGCTCAC
[0069] MFA / Prpos-F: AGGGTTTTTCCCAGTCACGACTGCCATTACCCAGGCCGACGCAGC
[0070] MFA / Prpos-R:CTTCTCCCCTTACCCCATAAGGTGGCTCCTACCCGTGATC
[0071] MFA / gfp-F:GTAGGAGCCACCTTATGGGTAAGGGAGAAGAACTTTTCAC
[007...
Embodiment 3
[0079] Example 3 Application of DATEL to quickly construct a combinatorial library of β-carotene synthesis pathway
[0080] In order to verify the assembly ability of multiple fragments of DATEL, the crtEXYIB of the β-carotene synthesis gene cluster derived from Pantoea agglomerans was used for assembly application. The gene cluster is divided into five DNA segments crtE, crtX, crtY, crtI and crtB (such as Figure 5 shown), and designed and assembled into the vector pUC19 according to this sequence. In particular, as a significant functional application of the DATEL scarless assembly technology (combinatorial assembly of modified proteins and mutations in metabolic pathways), the RBS sequence in each upstream forward primer was degenerately designed, and PCR amplified using this primer Upstream of each gene fragment is an RBS library with extremely rich diversity (protein translation intensity). The primer information of this embodiment is as follows:
[0081] CPA / puc19-F: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com