Rapid detection method for millet smut fungi
A detection method, millet technology, applied in the detection of millet smut infection and the field of rapid molecular detection of millet smut, can solve the problems of unfavorable disease transmission, time-consuming, labor-intensive, time-consuming and other problems, and achieve timely control of diseases Spread, simple operation, high specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Method for detection of millet smut pathogen by PCR technique:
[0019] 1. Design of PCR-specific primers: According to the difference between the ITS sequence of the pathogen of millet smut and the ITS sequence of other smut species on NCBI, PCR-specific primers were designed using the software Primer Premier 5.0, and the upstream primer F1 sequence was 5'-AGACGGGTTTACCACTCAAC -3', the downstream primer R1 sequence is 5'-AAGCCACGGTGAATGGCAAAG-3', and was synthesized by Bioengineering (Shanghai) Co., Ltd.
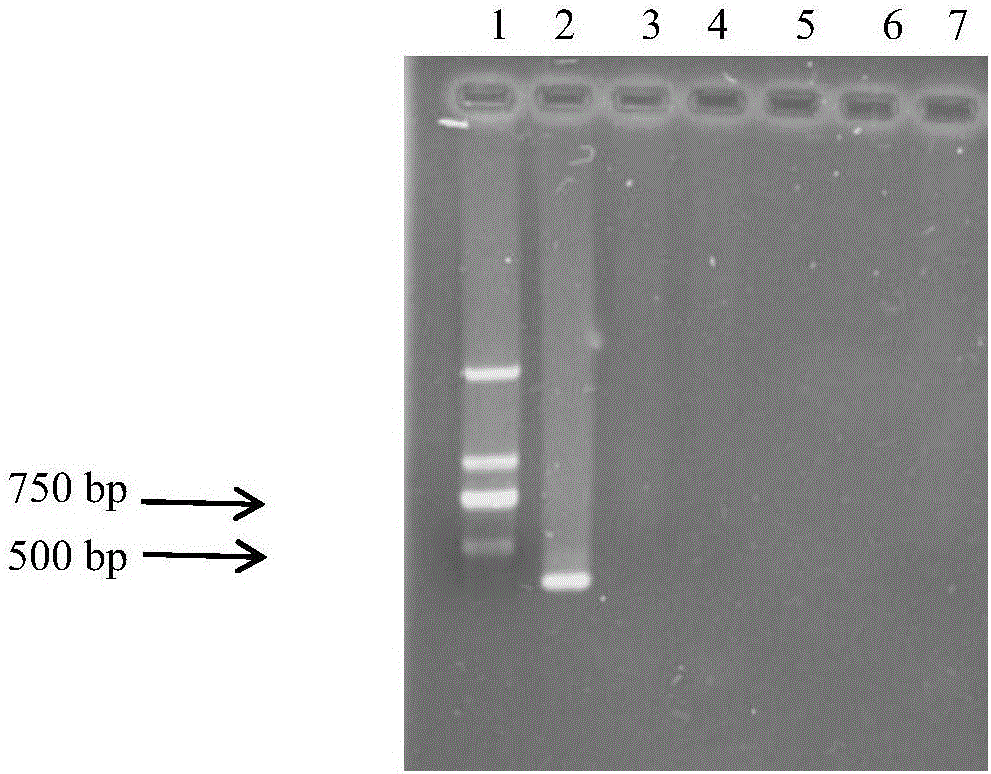
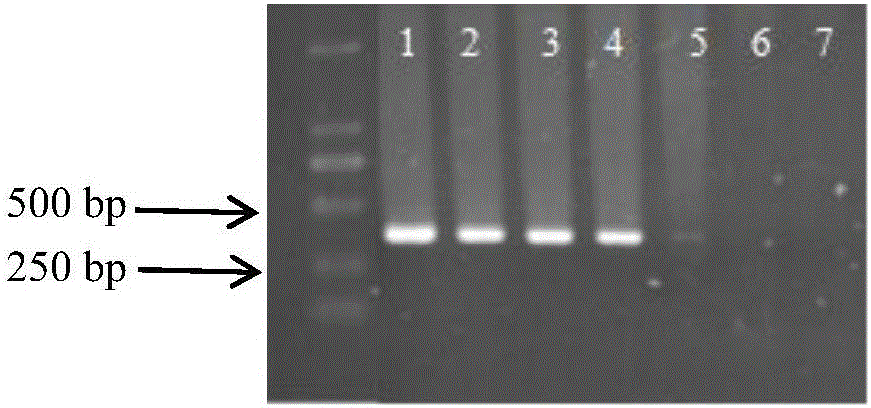
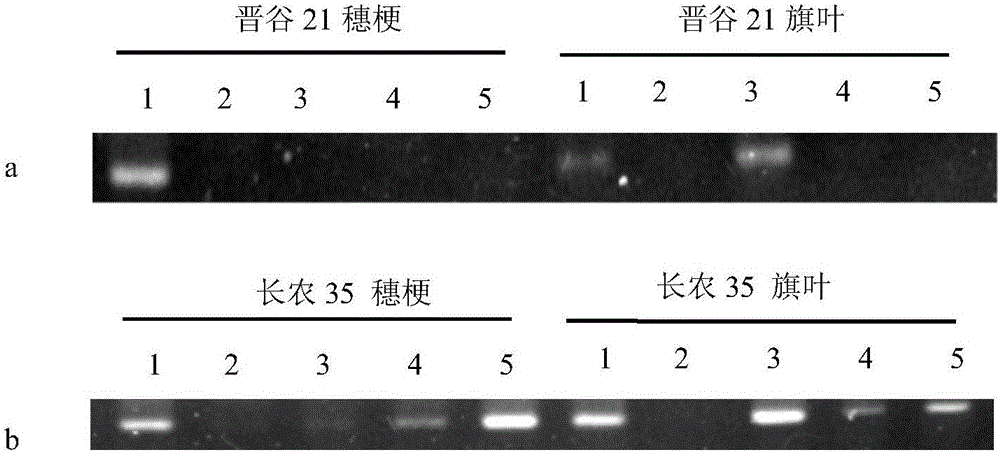
[0020] 2. Detection of primer specificity: the genomic DNA of the smut spores and the tested pathogenic bacteria and the millet leaves of the control group were subjected to primer specificity detection. The pathogenic bacteria tested included spores of white pathogen of millet, spores of corn smut, and hyphae of blast fungus.
[0021] The reaction system is 20 μL, including: 2 μL of 10×Buffer, 1.6 μL of dNTP, 0.4 μL of each primer, 0.4-1 μL of DNA template, 0.2 μL ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com