Primer group utilizing LAMP technology to detect witloof pseudomonas and kit and method
A technology for detection of Pseudomonas cichorii, which is applied in the directions of recombinant DNA technology, biochemical equipment and methods, and microbial determination/inspection, can solve the problems of long time for amplification and product detection, and achieves improved sensitivity and improved Specific, adaptable effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] The design of embodiment 1 primer
[0033] According to the specific region of the 16S rRNA gene (Genbank accession number KU923372) of Pseudomonas cichori that infects tomatoes in my country registered in Genbank, the outer primers F3 and B3 and the inner primers FIP and BIP were designed, and the following primer sets were finally determined:
[0034] F3: 5'-CAGCCGCGGTAATACAGAG-3'
[0035] B3: 5'-GCCACTGGTGTTCCTTCC-3'
[0036] FIP: 5'-GCCCGGGGATTTCACATCCAACGGTGCGAGCGTTAATCGG-3'
[0037] BIP: 5'-ACTGCATCCAAAACTGGCGAGCGCATTTCACCGCTACACAGG-3'.
Embodiment 2
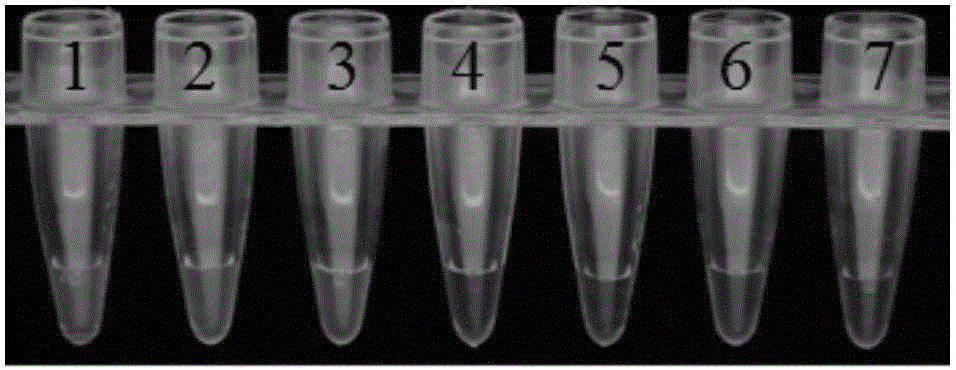
[0038] Example 2 Specific amplification of LAMP primers to Pseudomonas cichorium
[0039] 1. Experimental materials
[0040] With Pseudomonas chicory (strain number is Pc-Gd-3, this strain has been disclosed on GenBank, and the accession number of its 16Sr RNA is KU923372), solanacearum (strain number is SSf-4, this strain has been Disclosed on GenBank, the accession number of its 16SrRNA is FJ744124), Pectin bacillus black rot (the strain number is P1, the strain has been published on GenBank, the accession number of its 16SrRNA is KC695819), Pectobacterium carotovorum subsp.brasiliense (strain The serial number is Pcb-zj-1, and the 16Sr RNA sequence of this strain has been registered in the International Gene Bank (http: / / www.ncbi.nlm.nih.gov, accession number KX377593) as material.
[0041]The Pseudomonas cichorium LAMP detection kit includes the following components: the LAMP primer set designed in Example 1, the LAMP reaction solution, the enzyme solution and the nucleic...
Embodiment 3
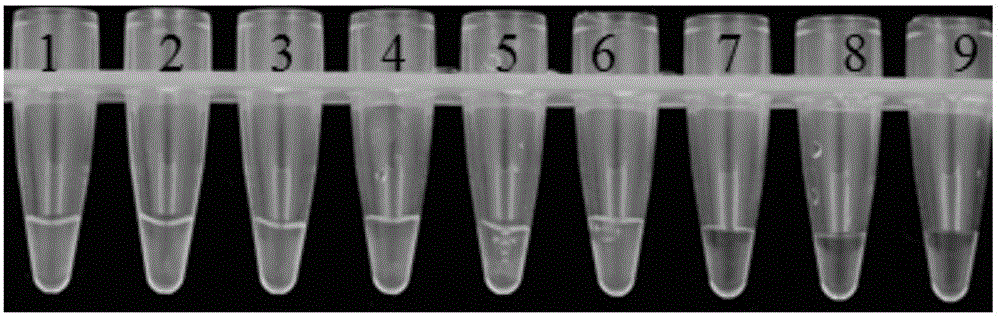
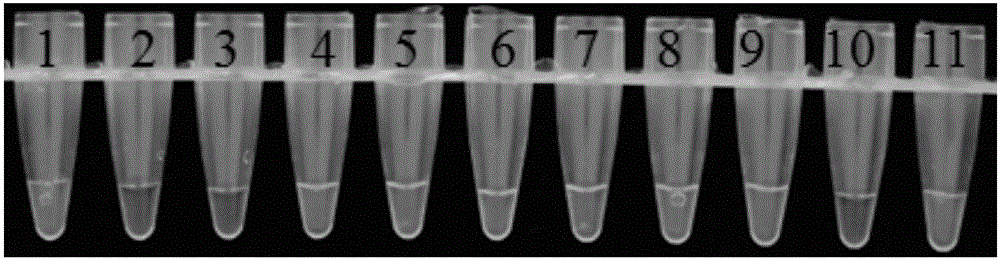
[0052] Example 3: Sensitivity determination for detection of Pseudomonas cichorium
[0053] 1. Experimental materials
[0054] Pseudomonas chicory as material.
[0055] Pseudomonas chicory LAMP detection kit is the same as in Example 2.
[0056] 2. Experimental method
[0057] (1) Genomic DNA extraction: the method is carried out according to the bacterial genomic DNA extraction kit;
[0058] (2) Take 10 μL of the above-mentioned genomic DNA in a PCR tube as the template mother solution, and dilute the template mother solution by 10 -1 , 10 -2 , 10 -3 , 10 -4 , 10 -5 , 10 -6 , 10 -7 times, take 1 μL from each serial dilution as a template;
[0059] (3) Prepare the loop-mediated isothermal amplification (LAMP) reaction mixture, including 1 μL of each gradient dilution DNA, 12.5 μL of 2×LAMP reaction solution, 3 μL of each primer FIP and BIP with a concentration of 20 μM, and 1 μL each of primers F3 and B3 with a concentration of 10 μM , Bst DN polymerase 1μL, Fluor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com