Method for detecting microRNA content by using single-stranded specific nuclease
An enzyme detection, single-strand technology, applied in the direction of DNA / RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of increased false positive rate of diagnostic results and reduced hybridization stringency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Embodiment 1, the method of using SSSN enzyme to detect whether the sample to be tested contains microRNA let-7a
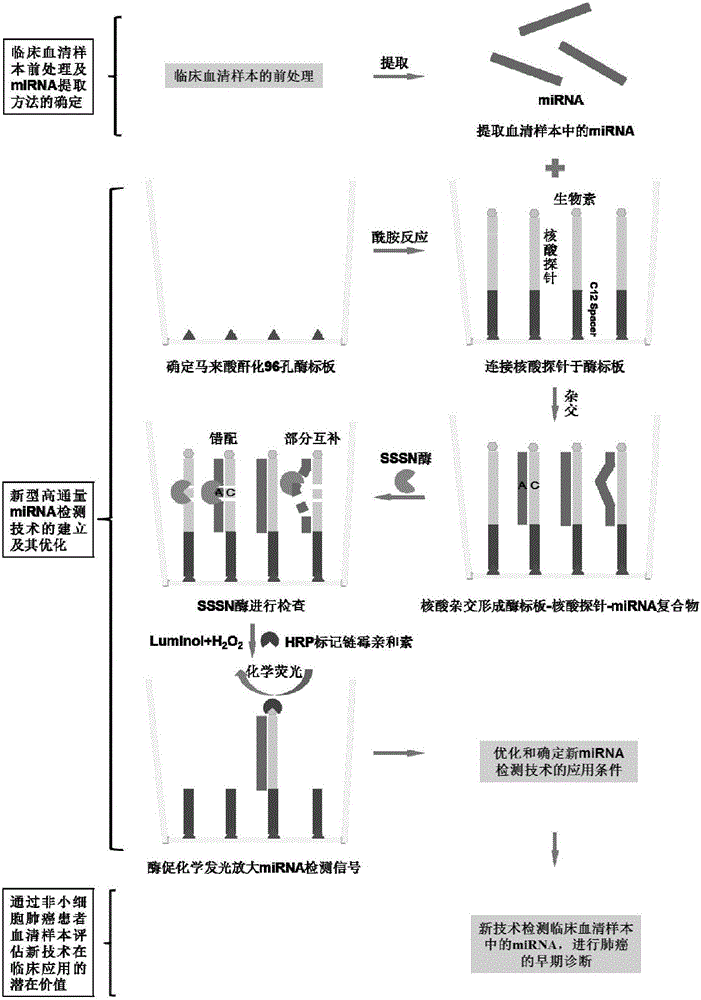
[0053] figure 1 It is a flowchart of microRNA detection using SSSN enzyme.
[0054] In the following examples, microRNA let-7a is used as the microRNA to be detected, and the nucleotide sequence of microRNA let-7a is UGAGGUAGUAGGUUGUAUAGUU (sequence 2).
[0055] 1. Samples to be tested
[0056] The embodiment of the present invention adopts the microRNA let-7a solution with a concentration of 0.5fM to 1000fM as the sample to be tested;
[0057] The solute in the above solution is microRNA let-7a, and the solvent is a PBS buffer solution (8g NaCl, 0.2g KCl, 1.44g Na 2 HPO 4 and 0.24g KH 2 PO 4 , dissolved in 1000ml of water).
[0058] The sample to be tested in the present invention can also be derived from plasma or serum, and the RNA of plasma or serum needs to be extracted. The specific method is as follows: the plasma or serum sample is diluted to...
Embodiment 2
[0089] Embodiment 2, the method that utilizes SSSN enzyme to detect microRNA let-7a content in the sample to be tested
[0090] 1. Samples and standards to be tested
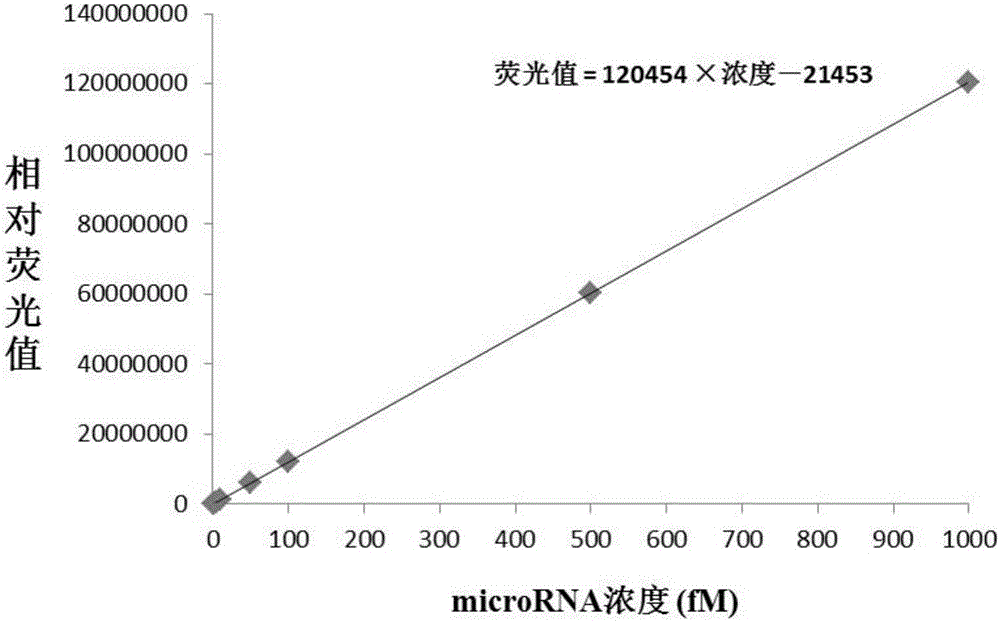
[0091] Standards with different concentrations are microRNAlet-7a solutions with concentrations of 0.5fM, 1fM, 5fM, 50fM, 500fM, and 1000fM;
[0092] The samples to be tested are microRNA let-7a solution with a concentration of 10fM and microRNA let-7a solution with a concentration of 100fM.
[0093]The solute in the above solution is microRNA let-7a, and the solvent is a PBS buffer solution (8g NaCl, 0.2g KCl, 1.44g Na 2 HPO 4 and 0.24g KH 2 PO 4 , dissolved in 1000ml of water).
[0094] 2. Method for high-throughput detection of microRNA let-7a content using SSSN enzyme
[0095] 1. MicroRNA let-7a probe preparation
[0096] Same as Example 1.
[0097] 2. Obtaining the microtiter plate after the reaction
[0098] 1) Microtiter plate for connecting probes
[0099] Same as Example 1.
[0100] 2) ELISA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com