Method for measuring activity of Phi29 DNA polymerase and rolling circle sequencing method
A polymerase and activity technology, applied in the field of determination of Phi29DNA polymerase activity, can solve problems such as difficult to achieve high throughput, automation, radioactive pollution, long cycle, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
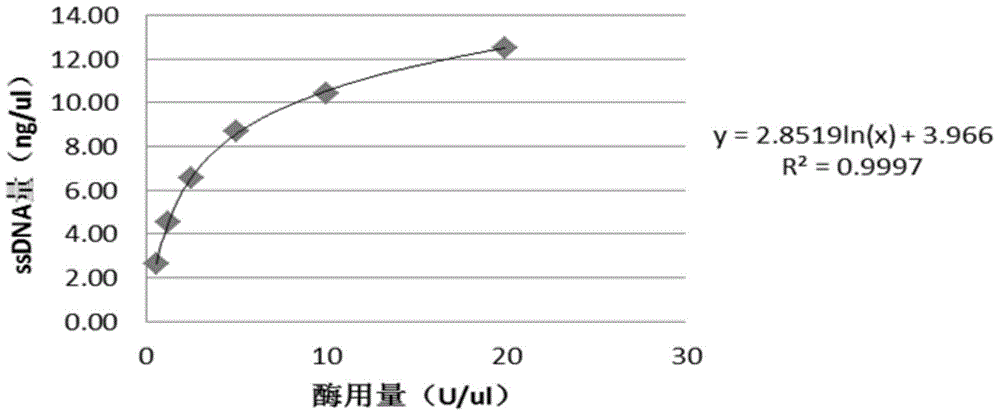
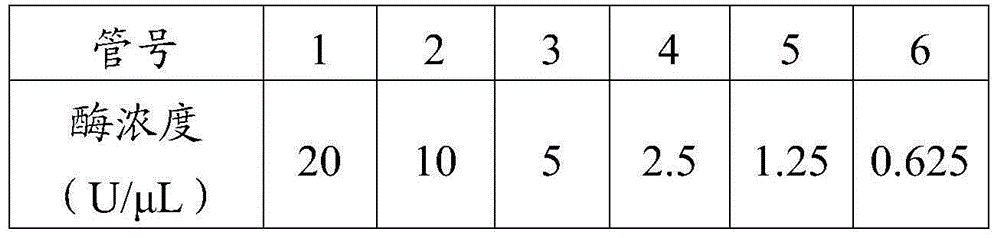
[0032] Example 1: Establishment of a method for measuring the polymerization activity of phi29 DNA polymerase by fluorescence method
[0033] 1. Standard curve establishment
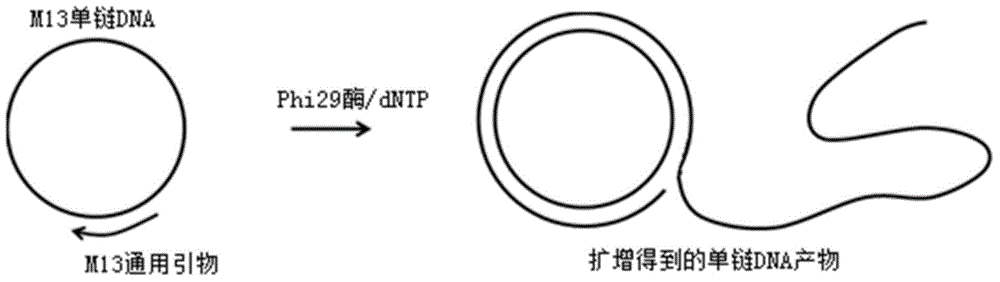
[0034] (1) Rolling circle amplification (RCA) reaction
[0035] a) The materials required for the RCA reaction are as follows:
[0036] Template: M13 single-stranded DNA (ssDNA);
[0037] Amplification primer: 5'-aagccatccgcaaaaatgacctct-3' (SEQ ID NO: 1);
[0038] Polymerase: Phi29DNA polymerase (Enzymatics).
[0039] b) RCA reaction steps:
[0040] i. Prepare the primer-template hybridization reaction mixture, the reaction system is shown in Table 1 (can be adjusted according to the actual situation), and the reaction mixture is placed in a PCR instrument for primer-template hybridization at 95°C for 3-5 minutes.
[0041] Table 1 Primer template hybridization reaction mixture reference system
[0042] components Dosage M13 ssDNA 1.5μL wxya 2 O water
83.3μL 10×phi2...
Embodiment 2
[0066] Example 2 Application of RCA products based on nanoarray optical sequencing platform
[0067] 1. For the RCA products obtained in Example 1, after taking a certain amount of RCA products for Qubit detection concentration, the remaining RCA products can be added to the chip of the nano-array, and high-throughput optical sequencing platform BGI-1000 is used for high-throughput sequencing. Throughput sequencing.
[0068] 2. Sequencing primer: complementary to the amplification primer of the RCA reaction. If the non-M13 single-stranded DNA used in Example 1 is a circular single-stranded DNA with a specific sequence, the sequence corresponding to the known specific sequence can be used as a sequencing primer.
[0069] 3. Specific process: preprocessing of samples to be tested—loading of samples to be tested—data generation of samples to be tested—data processing of samples to be tested—downstream application analysis.
[0070] 4. Application: The data obtained by sequencin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com