Biosynthetic gene cluster of Rubrolone and application of biosynthetic gene cluster
A technology for biosynthesis and roboluone, applied in the direction of plant genetic improvement, application, microorganism, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] 1. Sequencing and sequence bioinformatics analysis of the biosynthetic gene cluster of Roboloxone:
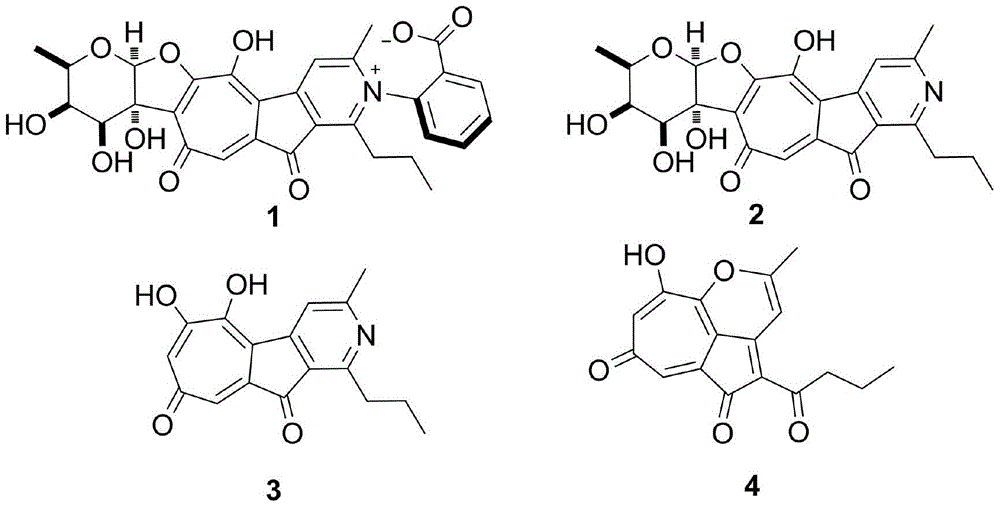
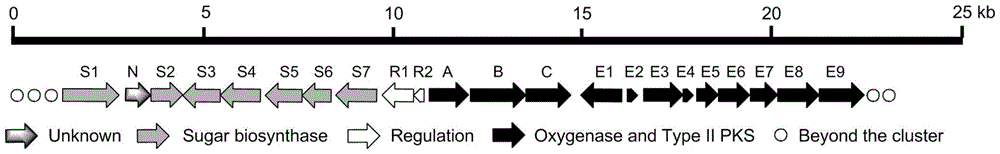
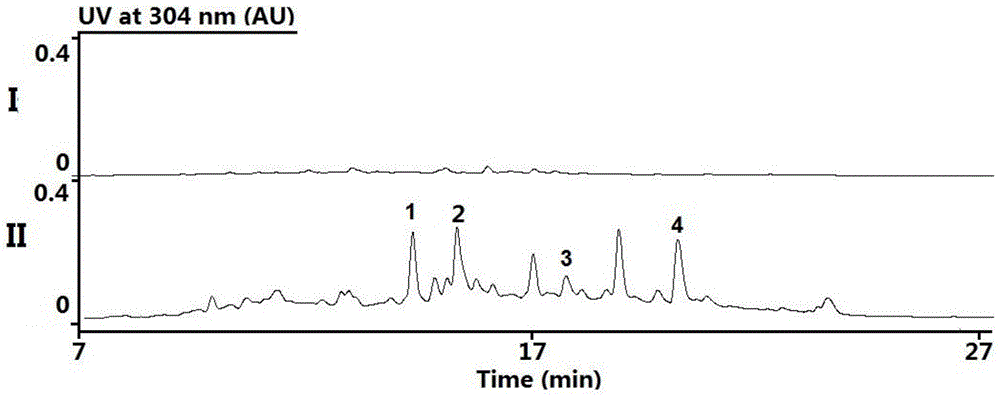
[0063] By sequencing the cosmid containing the robolutone biosynthetic gene cluster sequence as SEQ ID NO.1, the base sequence of the 6308th to 28758th position of the genome sequence SEQ ID NO.1 with an insert fragment of about 37Kb was obtained. Bioinformatics software and database comparison analysis showed that the sequenced Coase plasmid contained the entire Robolunone biosynthetic gene cluster, the gene cluster was about 22Kb in length, and analyzed that the gene cluster contained 24 open reading frames (open reading frames, ORFs), including genes responsible for the synthesis of the phenolic seven-membered ring skeleton of Roboloxone, namely rubA, rubB, rubC, rubE1, rubE2, rubE3, rubE4, rubE5, rubE6, rubE7, rubE8, rubE9, a total of 12 genes, responsible for The glycosyl synthesis genes of robolulone, i.e. rubS1, rubS2, rubS3, rubS4, rubS5, rubS6, rubS7, are 7 gene...
Embodiment 2
[0068] Streptomyces sp.CGMCC No.11535 (preserved on October 23, 2015 in the General Microbiology Center of China Committee for the Collection of Microbial Cultures, preservation address: No. 3, Yard 1, Beichen West Road, Chaoyang District, Beijing, Microbiological Research, Chinese Academy of Sciences from Kunming Institute of Botany, Chinese Academy of Sciences (No. KIB-H033).
[0069] Streptomyces sp.CGMCC No.11535 is a typical Streptomyces, its basal hyphae are branched, the aerial hyphae are slightly thicker, and some of the aerial hyphae differentiate into spiral sporocydus. At the same time, a red compound can be produced in the culture medium. The G+C content of its DNA is about 71%, which has typical characteristics of Streptomyces.
[0070] Robolone-producing bacteria Streptomyces Streptomyces sp.CGMCC No.11535 (KIB-H033) a large number of genomic DNA extraction:
[0071] Mycelia culture: use 50ml of TSB liquid medium to culture on a shaker at 28°C and 250rpm for 48...
Embodiment 3
[0073] Construction of Genome Library of Streptomyces sp. CGMCC No.11535 (KIB-H033) Genomic Library of Robolocone-producing Bacteria:
[0074] First, the amount of restriction endonuclease Mbo I was determined through a series of dilution experiments. In a 50 μL system, 5 μL of Streptomyces sp.CGMCC No.11535 genomic DNA, 5 μL of 10× reaction buffer and 7.5 μL Dilution of 10 -2 For Sau3A I, stop the reaction with 2.5 μL of 15 mM EDTA and the appropriate loading buffer. On this basis, a genomic DNA fragment slightly larger than 40kb was obtained by a large number of partial enzyme digestions, and dephosphorylated with a dephosphorylase.
[0075] The vector pJTU2554 plasmid used to construct the library was first cut from the middle of the two cos sequences with the restriction endonuclease Hpa I, then dephosphorylated, and then used the restriction endonuclease Bam HI from the multiple cloning site Cut open to obtain two arms. The treated vector was ligated with the previousl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com