Method for generating alfalfa genome edited homozygous plant
A genome editing and alfalfa technology, applied in the field of plant genetic engineering, can solve the problems of prolonging the test cycle and long plant growth cycle, and achieve the effects of simplifying the workload, growing rapidly, and shortening the screening cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Embodiment 1: CRISPR / Cas9-PDS expression vector construction and transformation obtain Medicago truncatula (R108) hairy root root system, which comprises the following steps:
[0050] 1) Cloning and gRNA design of Medicago truncatula (R108) PDS gene:
[0051] Cloning of PDS gene: Searched by NCBI and Phytozome, there is only one complete PDS gene sequence (gene number Medtr3g084830) in Medicago truncatula genome. The sequence of this gene was used as a standard to design primers, and the DNA of Medicago truncatula (R108) was used as a template for PCR amplification and sequencing.
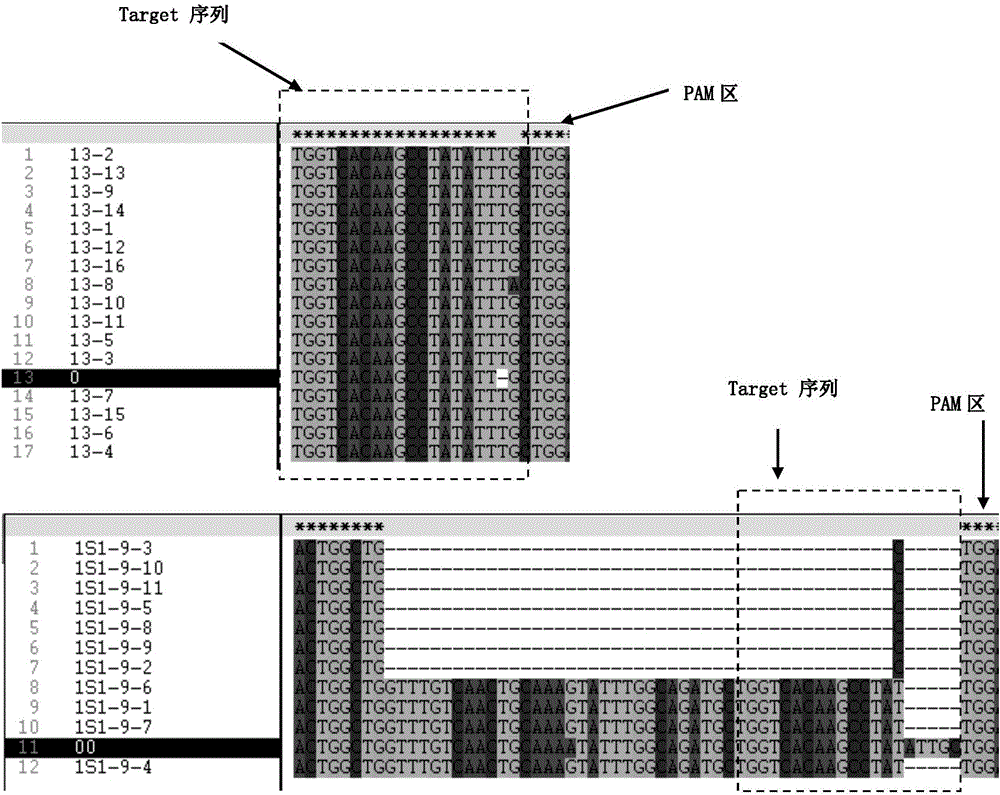
[0052] gRNA design: use CRICPR-P website, use Medicago truncatula (Mt4.0v2) as TargetGenome, use Medtr3g084830 gene as Locus Tag, Submit. Note that gRNA with suitable restriction sites adjacent to the PAM region is selected as the target site for site-directed editing.
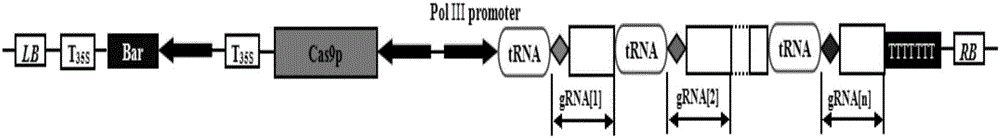
[0053] 2) Construction of genome editing expression vector pYLCRISPR / Cas9P35S-B-tRNA-PDS:
[0054] PUC57 vector plasmid...
Embodiment 2
[0073] Example 2: Rapid screening of homozygous hairy root systems where the PDS gene of Medicago truncatula (R108) was site-specifically edited. The main steps are as follows:
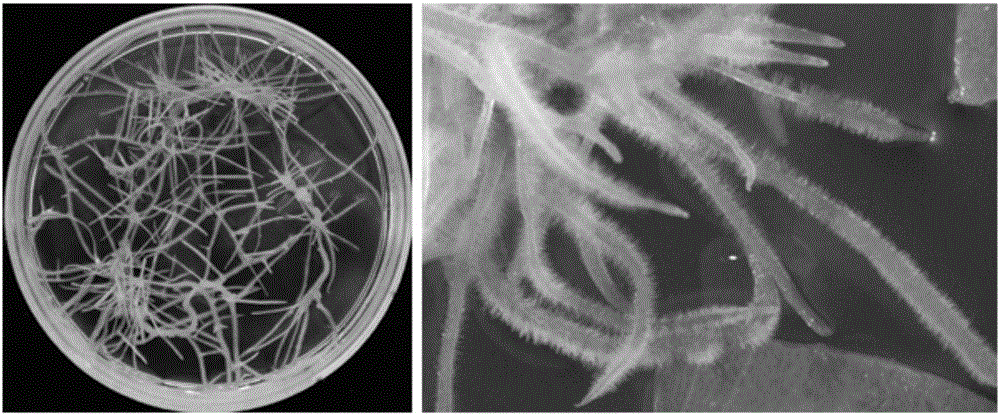
[0074] 1) Propagation of hairy roots: The resistant hairy roots induced in Example 1 were randomly numbered, and the root systems were randomly numbered 1S0, 2S0, 3S0... in order to facilitate tracking and purification. The resistant root system grows rapidly and can be subcultured every 10-14 days. In the S1 and S2 generations, 5-10 single hairy roots were randomly selected for continuous subculture, and the remaining root segments after subculture were collected for DNA extraction and reserved for identification. The temperature of the incubator was 25±2°C, and the culture was carried out in the dark.
[0075] 2) Enzyme digestion identification of hairy root: Extract DNA from the resistant hairy root sample obtained in step (1), PCR amplify the target gene including gRNA, and perform single enzyme ...
Embodiment 3
[0082]Example 3: Establishment of the hairy root regeneration system and the homozygous editing of the PDS gene to obtain the albino seedlings of Medicago truncatula (R108), the main features of which include the following steps:
[0083] 1) Hairy root induction of callus: Transfer the homozygous root system of genome editing identified by screening to callus induction medium (SHC), induce it for 3-5 weeks, and the callus produced can proceed to the next step of differentiation and regeneration. The temperature of the incubator was 25±2°C, and the culture was carried out in the dark.
[0084] 2) Hairy root differentiation and regeneration: the callus obtained in step (1) is transferred to the differentiation medium (MSR), and bud points can be differentiated within one week, and regenerated shoots can be obtained after 3-5 weeks of differentiation. The temperature of the incubator is 25±2°C, and the light time is 16h light / 8h dark every day (see Image 6 ).
[0085] 3) Rooti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com