Streptomyces virginiae IBL14 type I-B-sv14 type CAS gene editing system
An ibl14typei-b-sv14, gene editing technology, applied in the field of genetic engineering in biotechnology, to achieve the effect of effective gene editing and immunity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 (EC JM109 (DE3) β-galactosidase gene galM knockout)
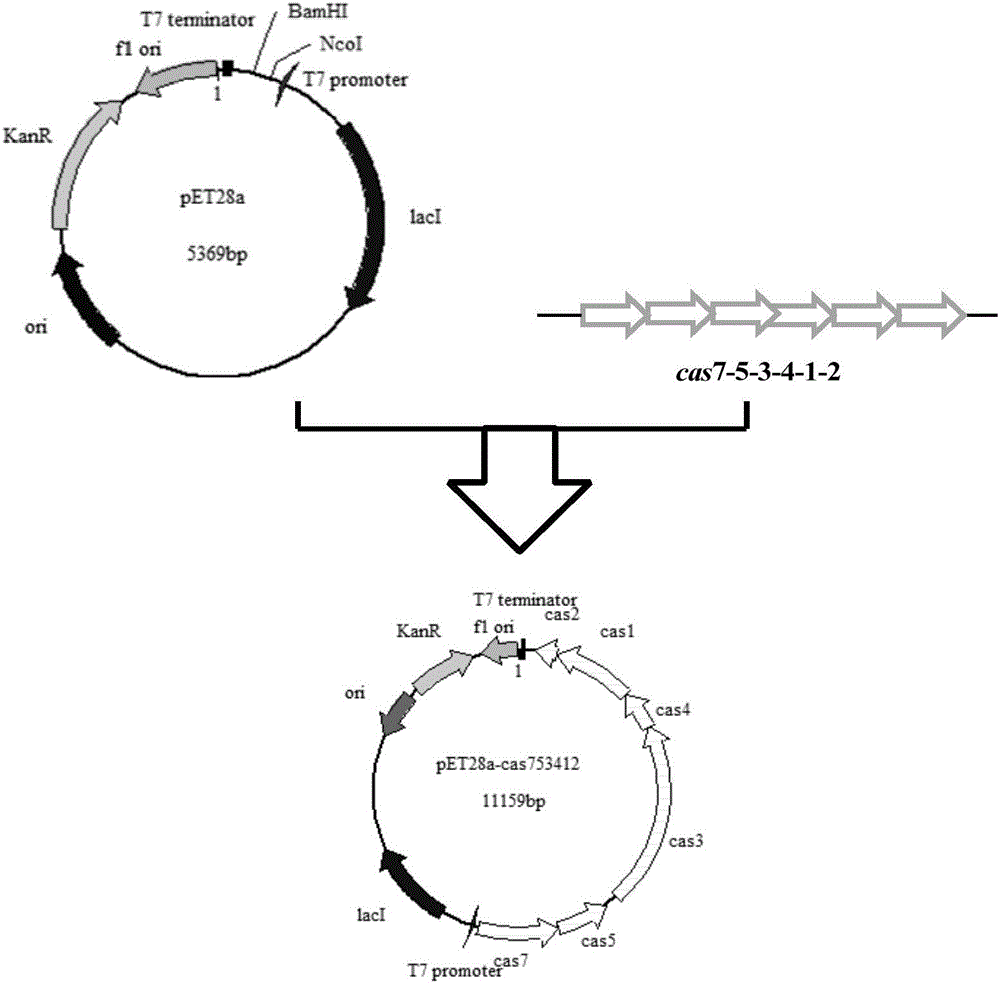
[0036] (1) Construction of Cas7-5-3-4-1-2 protein expression plasmid
[0037] According to the whole genome sequencing information of SV IBL14 and the sequence information of plasmid pET28a, the gene cas7-5-3-4-1-2 specific primers cas7-5-3-4-1-2-F and cas7-5-3-4 were designed -1-2-R (table 2); extract SV IBL-14 genomic DNA, use the DNA polymerase (TransTaq DNA Polymerase HighFidelity) produced by Beijing Quanshijin Biotechnology Co., Ltd. to carry out cas gene PCR amplification, reaction condition: 95 2min at ℃, 20s at 95℃, 20s at 60℃, 3min at 72℃, 250units of TransTaq DNA Polymerase High Fidelity (50μl reaction system), 30 cycles, 10min at 72℃. The PCR product was detected by 1% agarose electrophoresis, the kit was recovered, and the purified cas full-length gene fragment was obtained; the full-length cas gene sequence was connected to the plasmid pET28a by one-step method to obtain the protein expressio...
Embodiment 2
[0054] Cm in embodiment 2 (EC JM109 (DE3) R Insertion of resistance gene)
[0055] (1) Protein expression plasmid pET28a-cas7-5-3-4-1-2
[0056] With embodiment 1 step (1)
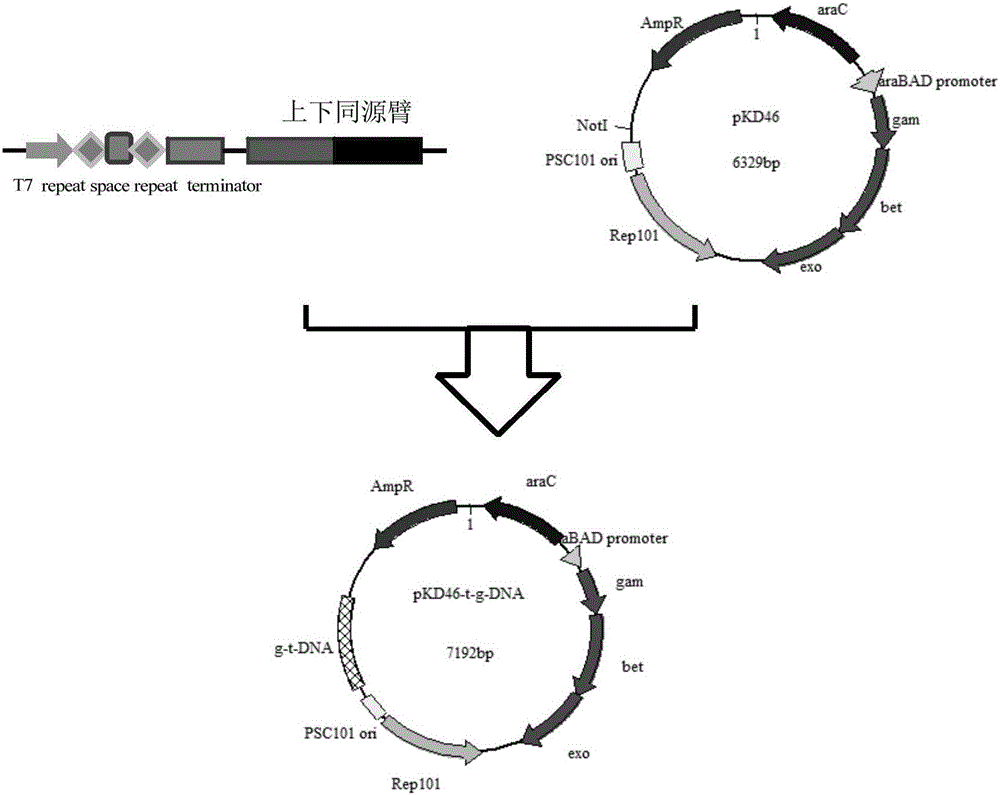
[0057] (2) Construction of gene editing plasmid pKD46-t / g-DNA
[0058] Divided by chloramphenicol (Cm R ) The plasmid pKD3 of the resistance marker gene is used as a template, combined with the upstream and downstream homology arms in the galM gene knockout, the PCR reaction is carried out to synthesize the upstream and downstream homology arms and insert chloramphenicol (Cm R ) The PCR product of the resistance marker was connected to the plasmid pKD46 to form the gene editing plasmid pKD46-galM-cm-t / g-DNA; the rest of the steps were the same as step (2) in Example 1.
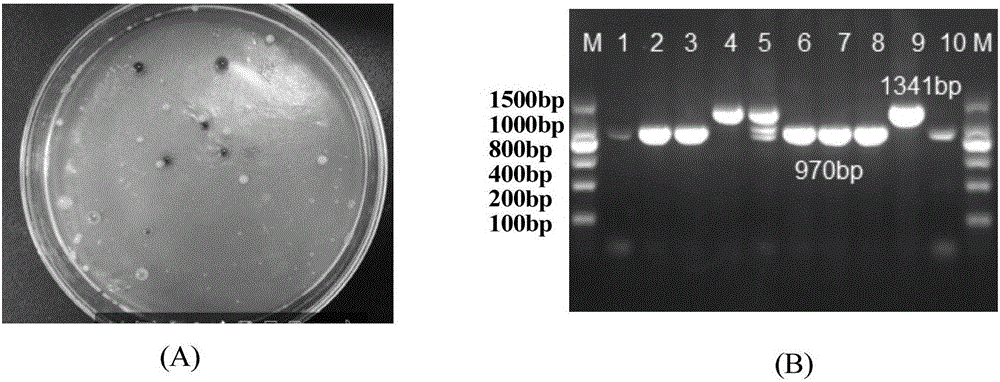
[0059] (3) Acquisition and inspection of recombinants
[0060] After induction with the obtained recombinant strains, transfer to TB liquid medium and culture at 39°C for 15 hours, then dilute and spread the obtained bacterial liquid,...
Embodiment 3
[0061] Example 3 (Immunization of EC JM109 (DE3) to Plasmid pKD3-Repeat Sequence in EC JM109 (DE3))
[0062] (1) Construction of protein expression plasmid pET28a-cas7-5-3-4-1-2
[0063] With embodiment 1 step (1)
[0064] (2) Gene editing plasmid pKD46-galM-pKD3-g / t EC - Construction of DNA
[0065] According to the DNA gene sequence in the plasmid pKD3, NotI enzyme cutting sites were directly synthesized at the beginning and end respectively, and the promoter T7, spacer (pKD3 base sequence), repeat (the repeat sequence in the CRISPR of EC JM109 (DE3)) and terminator pKD3-t EC -DNA; the galM-g-DNA in step (2) of Example 1 is connected to the plasmid pKD46 by T4 ligase to obtain the gene editing plasmid gene editing plasmid pKD46-galM-pKD3-g / t EC -DNA; the rest of the steps are the same as step (2) in Example 1.
[0066] (3) Acquisition and inspection of recombinants
[0067] Same as step (3) of Example 1; the recombinant ECJM109(DE3)-galM-pKD3 was obtained after gene ed...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com