Primers capable of simultaneously detecting various diarrhea pathogenic bacteria and application of primers
A technology of pathogenic bacteria and pathogenic bacteria, applied in the field of kits for simultaneous detection of multiple diarrhea pathogenic bacteria, can solve problems such as cumbersome operation, missed detection, and large human influence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
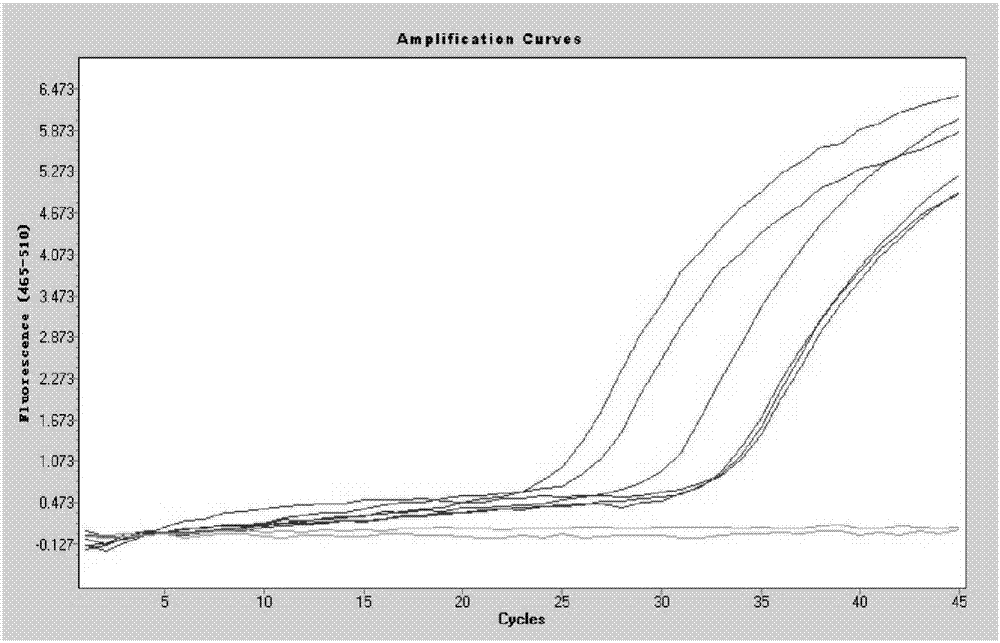
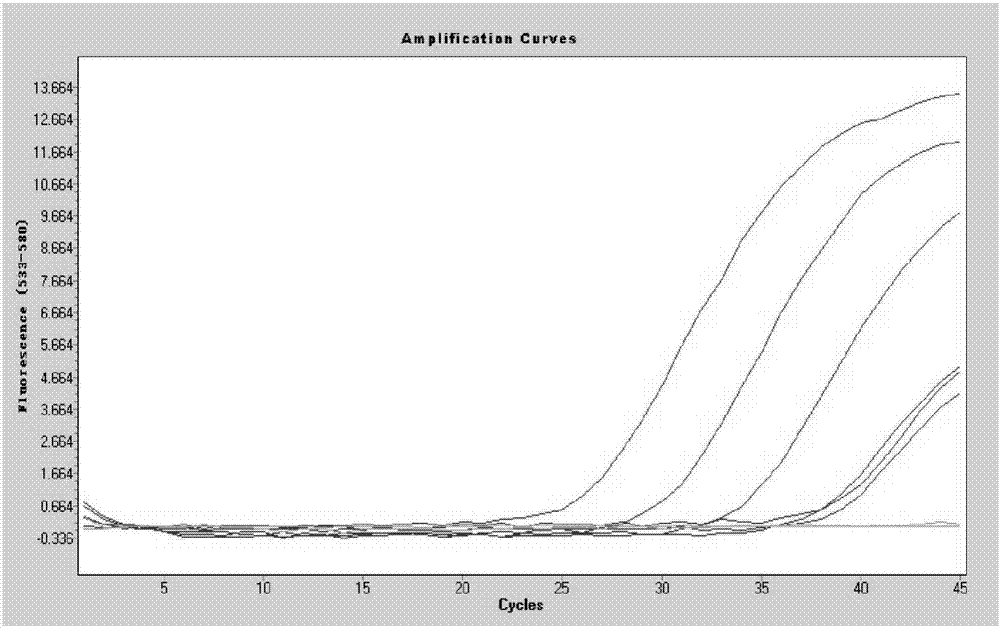
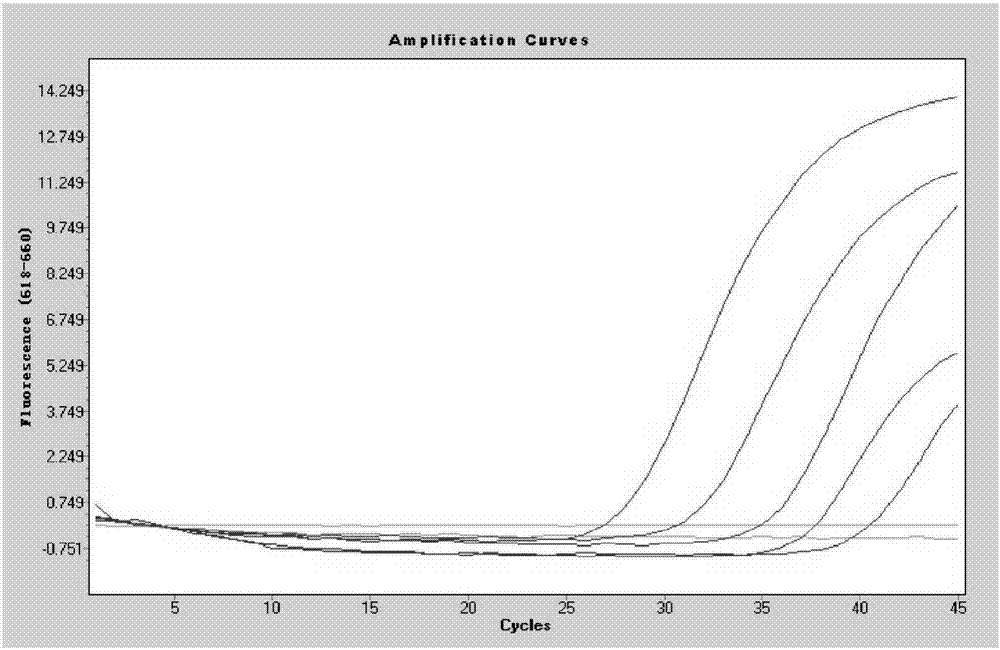
Image
Examples
Embodiment 1
[0098] The primers and probes for detecting 13 kinds of diarrhea pathogens were synthesized by Yingwei Jieji (Shanghai) Trading Co., Ltd. In practical application, the primer pairs and probes of three kinds of bacteria are divided into one group, as follows:
[0099] The first set of primer pairs for amplifying the nuc gene of Staphylococcus aureus, the ipaH gene of Shigella, and the tlh gene of Vibrio Parahemolyticus includes the following three pairs of primers and probes:
[0100] Primer pairs and probes for detection of S. aureus nuc gene:
[0101] F1 (SEQ ID NO.: 1): ATCCTAAAAAAGGTGTAGAG,
[0102] R1 (SEQ ID NO.: 2): TATCAGTTCTTTGACCTTTG,
[0103] P1 (SEQ ID NO.: 3): FAM-ATATGGTCCTGAAGCAAGTGCA-MGB;
[0104] Primers and probes for detection of Shigella ipaH gene:
[0105] F2 (SEQ ID NO.: 4): ATAAAGTCAGAACTCTCC,
[0106] R2 (SEQ ID NO.: 5): AACGCATTTCCTTCACGG,
[0107] P2 (SEQ ID NO.: 6): VIC-ATGAGATAGAAGTCTACCTGGCCTTCC-TAMRA;
[0108] Primers and probes for detecting...
Embodiment 2
[0164] Embodiment 2: the preparation method of kit.
[0165] (1) PCR reaction solution: 10*PCR reaction buffer, Q-solution, DNA polymerase, Mg2+ and dNTPs, stored at -20°C;
[0166] (2) Primer and probe mixture: The nucleotide sequences shown in SEQ ID NO.: 1-39 were synthesized by Yingwei Jieji (Shanghai) Trading Co., Ltd., and then according to grouping, three primer pairs and probes were synthesized. The needles were mixed in one tube, dissolved in double distilled water, the final concentration of each primer was 1 μmol / L, and stored at -20°C;
[0167] (3) Positive control: containing 13 kinds of diarrhea pathogenic bacterial genomic DNA respectively, wherein, the concentration of each bacterial genomic DNA containing a drug-resistant gene is 10 ng / μL, and stored at -20°C;
[0168] (4) Negative control: the concentration of Escherichia coli genomic DNA was 100 ng / μL, and stored at -20°C.
Embodiment 3
[0169] Embodiment 3: detection method.
[0170] Instrument: Roche 480 fluorescent quantitative PCR detector, BECKMAN 22R desktop micro-refrigerated centrifuge, Eppendorf 5810R desktop refrigerated centrifuge, Taicang Hualida Laboratory Equipment Company WH-866 vortex oscillator.
[0171] (1) Fecal sample pretreatment: 1. Sample pretreatment: Weigh 1g of fecal sample and suspend it in 9ml sterile PBS, shake vigorously for 15min, centrifuge at 200r / min for 3 times, each time for 5min, collect the supernatant; then 5000r / min Centrifuge for 3 minutes, collect the bacterial pellet, suspend the pellet with 1ml PBS, and repeat the centrifugation until the supernatant is basically clear; finally, suspend the collected pellet with 1ml PBS, and store it in a 2ml Eppendorf tube at -20°C for later use.
[0172] (2) Preparation of bacterial genomic DNA template: referring to published literature, different types of bacterial specimens were used, and corresponding commercial genomic DNA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com