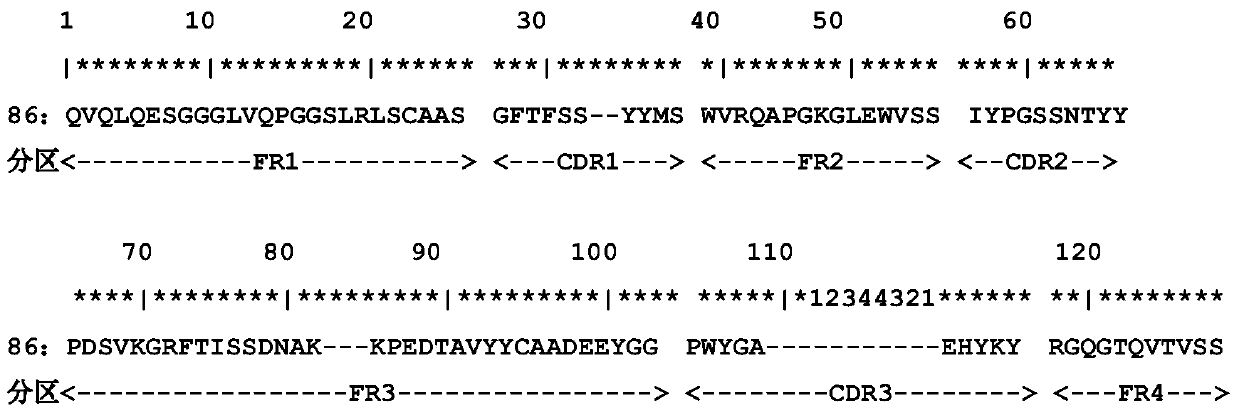
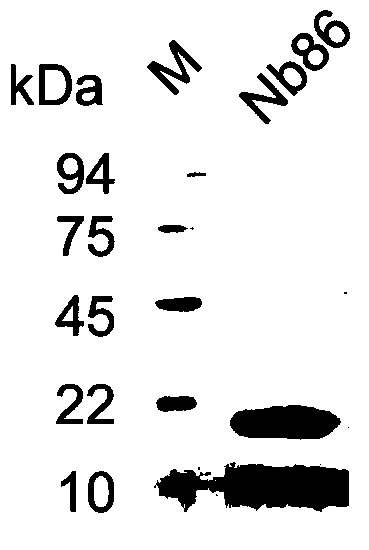
A CD105 nanobody nb86
A technology of nanobodies and DNA molecules, applied in the field of biomedicine or biopharmaceuticals, can solve the problems of lack of nanobodies and achieve the effect of good specificity and high affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Construction of a Nanobody library for CD105:
[0027] (1) First synthesize CD105 polypeptide, mix 1 mg CD105 with Freund's adjuvant in equal volume, immunize a Xinjiang dromedary, once a week, immunize 7 times in total, and stimulate B cells to express antigen-specific nanobodies;
[0028] (2) After the 7 times of immunization, extract 100mL camel peripheral blood lymphocytes and extract total RNA;
[0029] (3) Synthesize cDNA and amplify VHH by nested PCR;
[0030] (4) Use restriction enzymes PstI and NotI to digest 20ug pComb3 phage display vector (supplied by Biovector China Plasmid Vector Strain Cell Gene Collection Center) and 10ug VHH and connect the two fragments; (5) Transform the ligated product into electroporation In state cell TG1, the CD105 nanobody library was constructed and the storage capacity was determined, and the storage capacity was 1.85×10 8 .
Embodiment 2
[0031] Example 2: Nanobody screening process against CD105:
[0032] (1) Dissolve in 100mM NaHCO 3 , 20ug CD105 in pH 8.2 was coupled to a NUNC microtiter plate, and placed overnight at 4°C;
[0033] (2) Add 100uL 0.1% casein the next day, and block at room temperature for 1-3h;
[0034] (3) After 1-3h, add 100uL phage (5×10 11 tfu immunized camel nanobody phage display gene library), at room temperature for 1-2h;
[0035] (4) Wash 4-6 times with 0.05% PBS+Tween-20 to wash off unbound phages;
[0036] (5) Use 100mM TEA (triethylamine) to dissociate the phage that specifically binds to CD105, and infect Escherichia coli TG1 in logarithmic phase growth, culture at 37°C for 1 hour, produce and purify the phage for the next round For screening, the same screening process was repeated for 3-5 rounds to gradually obtain enrichment.
Embodiment 3
[0037] Embodiment 3: use the enzyme-linked immunosorbent method (ELISA) of phage to screen specificity single positive clone:
[0038] (1) From the cell culture dish containing phage after the above 3-5 rounds of selection, pick 96 single colonies and inoculate them in TB medium containing 100 micrograms per milliliter of ampicillin (1 liter of TB medium contains 2.3 grams of phosphoric acid Potassium dihydrogen, 12.52 grams of dipotassium hydrogen phosphate, 12 grams of peptone, 24 grams of yeast extract, 4 milliliters of glycerol), after growing to the logarithmic phase, add isopropylthiogalactoside ( IPTG), cultivate overnight at 30-35°C.
[0039] (2) Use the infiltration method to obtain the crudely extracted antibody, and transfer the antibody to an antigen-coated ELISA plate, and place it at room temperature for 1-1.5 hours.
[0040] (3) Wash off unbound antibodies with PBST, add primary mouse anti-HA tagantibody (purchased from Beijing Kangwei Century Biotechnology Co., ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com