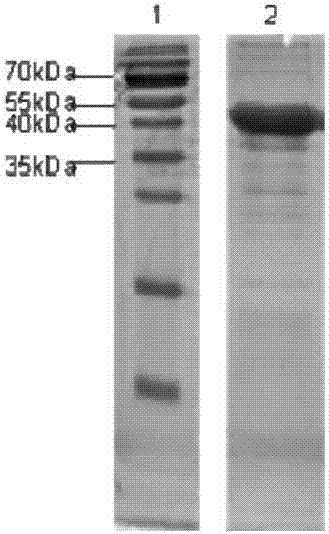
MRNA-coding nanobody and application thereof
A nanobody, coding technology, applied in applications, recombinant DNA technology, antibody mimics/scaffolds, etc., can solve problems such as destruction, cell transformation to produce cancer, and cell function destruction.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0113] Example 1: Expression and purification of HDAC6-CAT1 truncated protein:
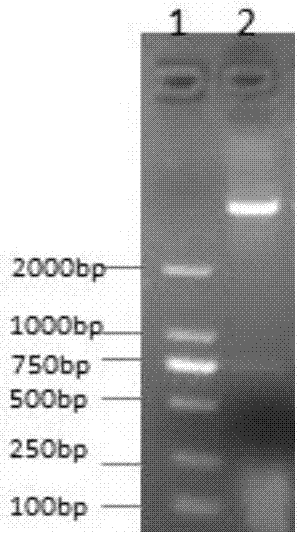
[0114] (1) According to the HDAC6 gene sequence, use Premier Primer5.0 software to design PCR primers: CAT1-JD-5-sal1 (cgaGTCGACgagcagttaaatgaattccattg) and CAT1-JD-3-not1 (gcgGCGGCCGCggcggccatctcacccttggggtcc), the length of the amplified gene fragment is 801bp; ( 2) Using CAT1-JD-5-sal1 and CAT1-JD-3-not1 as primers and HDAC6-WT recombinant plasmid as a template, 6-272 amino acids of HDAC6-Cat1 were amplified. 25 μL of PCR reaction system, containing 12.5 μL of 2× PCR Mix (containing enzyme), 1 μL of upstream primer, 1 μL of downstream primer, 1 μL of DNA template, and 9.5 μL of double distilled water. The cycle parameters of PCR were: pre-denaturation at 95°C for 8min; denaturation at 95°C for 40s, annealing at 57°C for 40s, extension at 72°C for 50s, 35 cycles; final extension at 72°C for 10min. 1% agarose gel electrophoresis observation. (3) Construct the pET32a-Cat1-JD recombinant plasmid,...
Embodiment 2
[0115] Example 2: Construction of the HDAC6-CAT1 nanobody library:
[0116] (1) Mix 1 mg of CAT1-JD recombinant protein antigen with Freund's adjuvant in equal volumes, and immunize a Xinjiang Bactrian camel once a week for a total of 7 consecutive immunizations. During the immunization process, B cells are stimulated to express specific nanobodies (2) After 7 times of immunization, extract 100ml of camel peripheral blood lymphocytes, and carry out ELISA antibody level detection of immunized camels, the results are as follows: image 3 As shown, the serum titer after immunization reached 10 4 , indicating that the HDAC6-CAT1-JD recombinant protein has a better immune effect; and extract total RNA; (3) synthesize cDNA and amplify VHH by nested PCR; (4) digest 20ug pMECS phage with restriction enzymes PstⅠ and NotⅠ Display the vector and 10ug VHH and connect the two fragments; (5) Transform the ligation product into electroporation competent cells TG1, construct the human antib...
Embodiment 3
[0117] Embodiment 3: Nanobody screening process against CAT1-JD recombinant protein:
[0118] (1) Take 200uL of recombinant TG1 cells and culture them in 2×TY medium, add 40uL of helper phage VCSM13 to infect TG1 cells during the period, and culture overnight to amplify the phages, use PEG / NaCl to precipitate the phages the next day, and centrifuge to collect the amplified phages ; (2) Couple 200ug of CAT1-JD recombinant protein dissolved in 100mM pH 8.2 NaHCO3 to a microtiter plate, place it overnight at 4°C, and set up a negative control at the same time; (3) Add 100ul of 3% BSA the next day, Block at room temperature for 2h; (4) After 2h, add 100ul amplified phage (2×10 11 tfu immunized camel nanobody phage display gene library), and reacted at room temperature for 1h; (5) Washed 5 times with PBS+0.05%Tween-20 to wash off the bound phage; (6) Use trypsin with a final concentration of 25mg / ml The phage specifically bound to the Fc fragment of the human antibody will be diss...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com