Method and device for detecting instability of microsatellite sites
A technology of microsatellite sites and instability, applied in the field of bioinformatics, can solve the problems of false positives and false negatives, and achieve the effect of increasing the number of detected sites and reducing the probability of false positives and false negatives.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
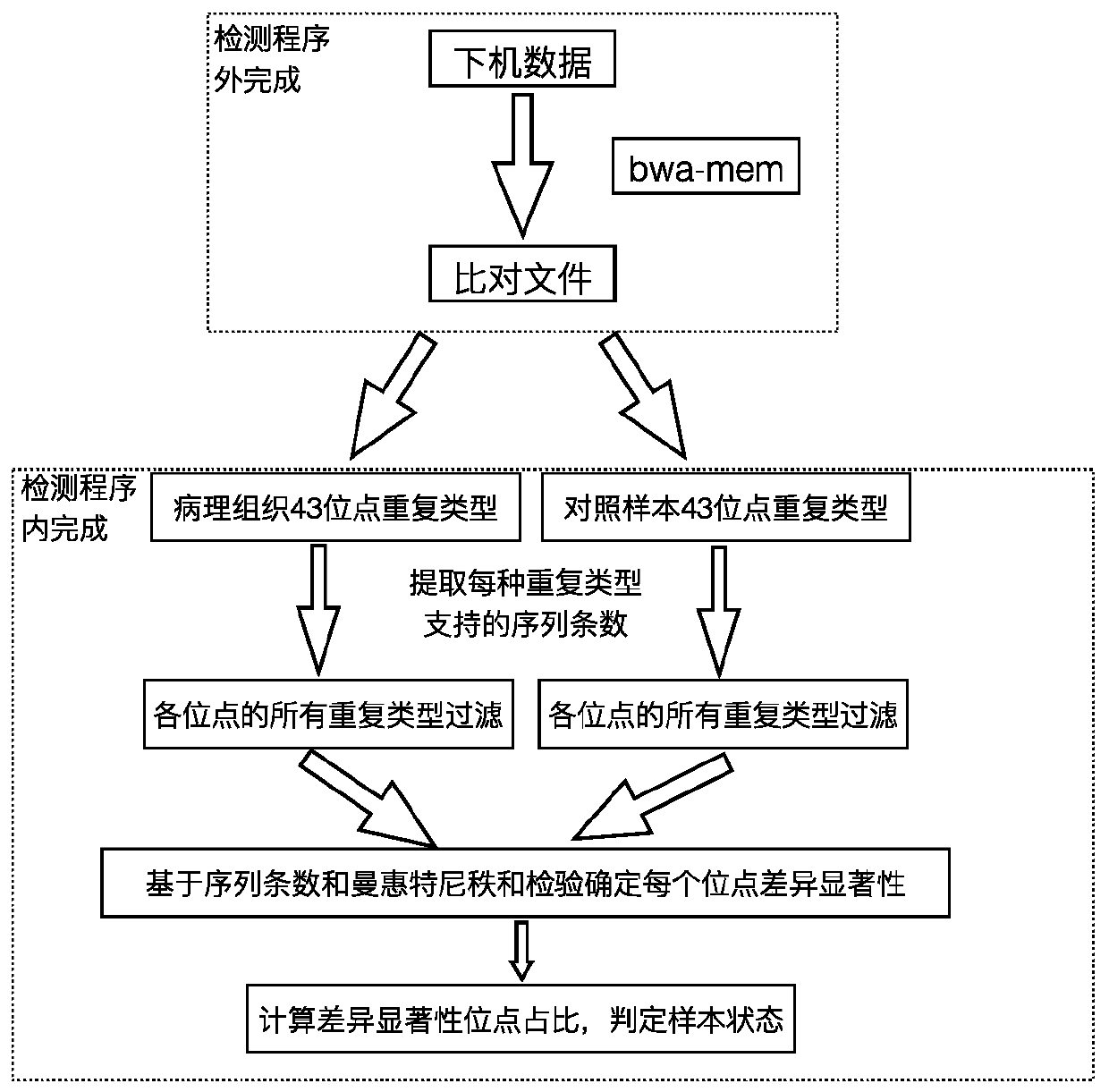
[0025] According to a typical embodiment of the present invention, the method is used to detect the instability of microsatellite sites in colorectal cancer.
[0026] The inventors of the present invention have screened out 43 microsatellite loci that are highly correlated with colorectal cancer, using these loci to judge the difference between pathological samples and control samples, and add statistical tests to determine the microsatellite instability of the samples , with high sensitivity and high specificity.
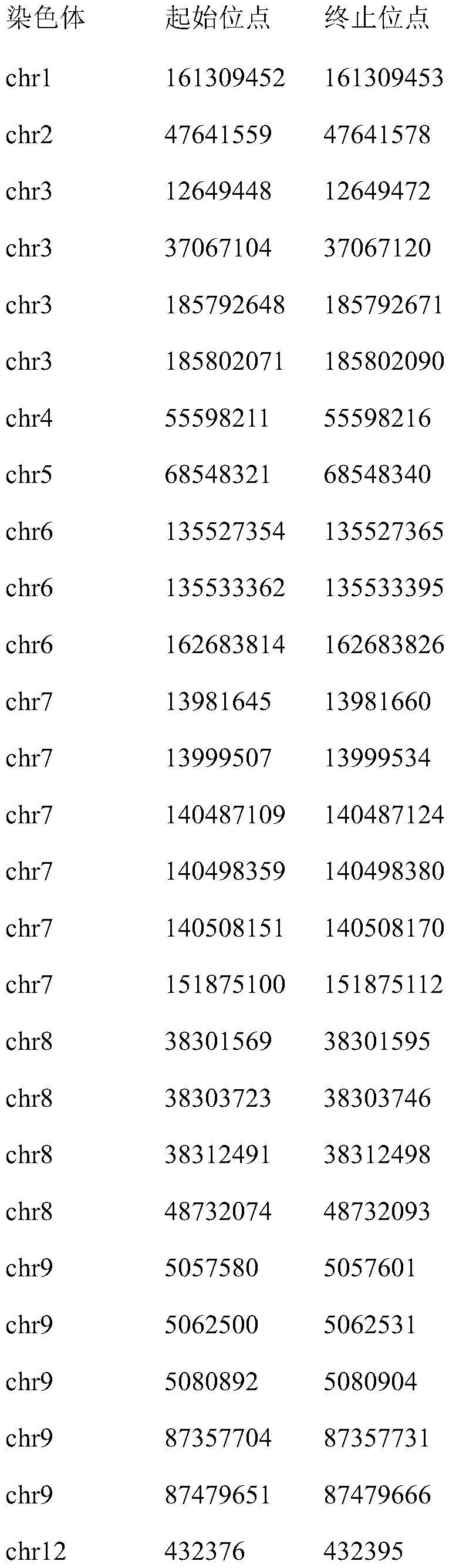
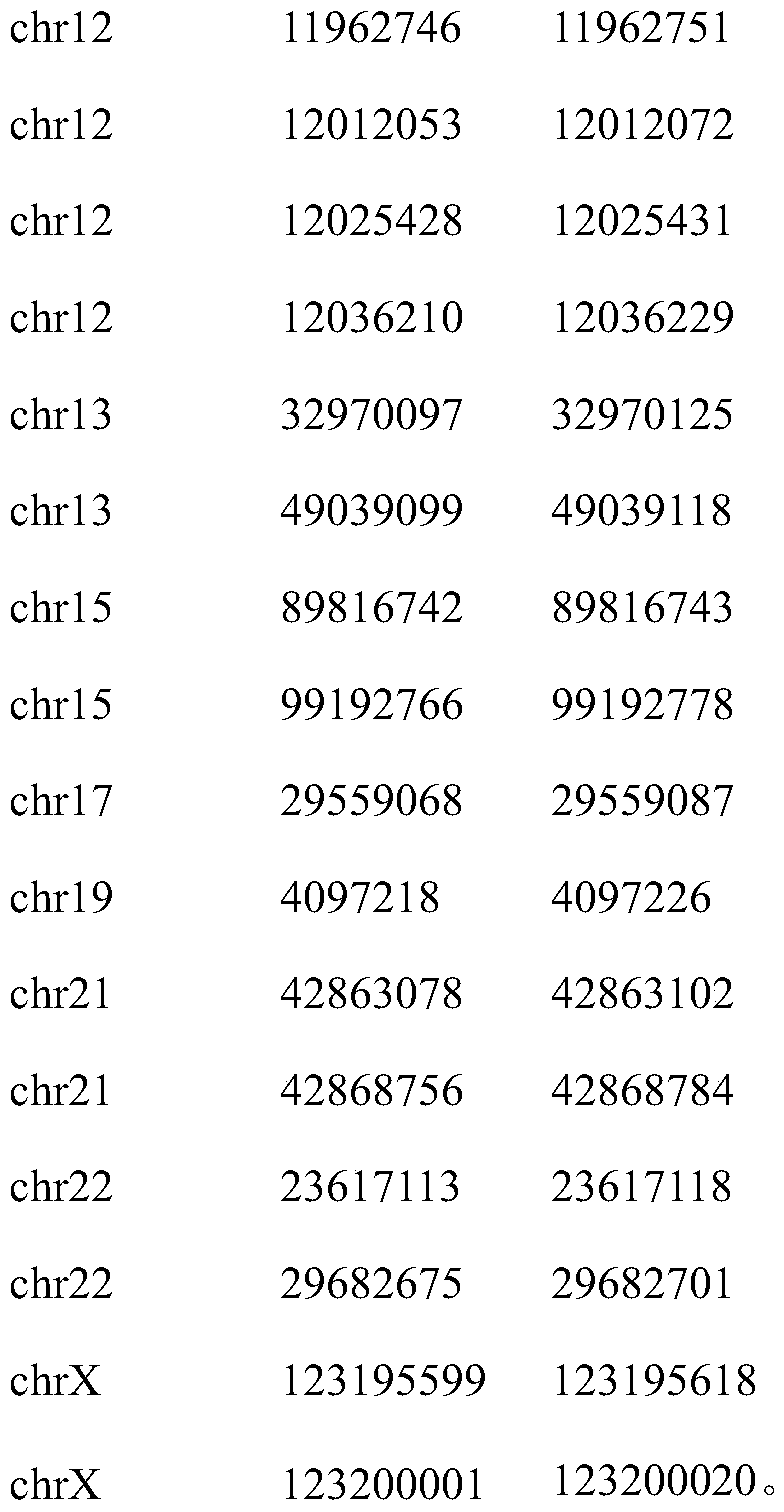
[0027] Regions of 43 microsatellite loci highly associated with colorectal cancer include:
[0028]
[0029]
[0030]
[0031] Preferably, the sample DNA includes pathological tissue sample DNA and control sample blood white blood cell DNA.
[0032] According to a typical implementation of the present invention, combined with the target region capture technology, high-throughput sequencing technology, combined with the determined information of 43 microsa...
Embodiment 1
[0043] In the first part of this embodiment, colorectal cancer pathological samples with known stable microsatellite sites and corresponding control samples are to be tested.
[0044] In the embodiment of the invention, the main reagent supplies are commercially available, and the information is as follows:
[0045]
[0046]
[0047] In this embodiment, the operation steps are as follows:
[0048] 1. Extract DNA (pathological tissue sample DNA and control sample blood leukocyte DNA), and use a fluorescence quantitator (Qubit) to quantify, the concentration is 3.8ng / ul, and the volume is 130ul; Make the DNA fragment size between 200-400bp, and then use agarose gel electrophoresis to detect whether the fragment size meets the requirements.
[0049] 2. Purify the fragmented DNA sample with magnetic beads first, and then perform end repair and 3' end adenylation. The system configuration is shown in Table 1. The basic steps are as follows: first incubate at 20°C for 30 minu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com