Reagent for detecting myocardial infarction and application of reagent
A technique for myocardial infarction and reagents, applied in the field of disease diagnosis, can solve the problems of limitation and atypical early clinical symptoms of AMI patients
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] The collection of embodiment 1 sample and total RNA extraction
[0047] The peripheral blood of patients with myocardial infarction and 6 cases of healthy controls were collected in the hospital from March 2015 to September 2016. Diagnostic criteria for acute myocardial infarction: developed according to the diagnostic criteria recommended by the third "Global Unified Definition of Myocardial Infarction" in 2012. An increase and / or decrease in the level of myocardial necrosis markers (mainly troponin) was detected, exceeding the 99th percentile value of the upper limit of the reference value at least once, and accompanied by at least one of the following ischemic symptoms.
[0048] 1) Symptoms of myocardial ischemia;
[0049] 2) New ischemic ECG changes;
[0050] 3) Pathological Q waves appear in the electrocardiogram;
[0051] 4) Imaging evidence shows new loss of myocardial activity or new local wall motion abnormality;
[0052] 5) Coronary angiography or autopsy ...
Embodiment 2
[0054] Example 2 Sequencing, data analysis and electronic verification
[0055] Sequencing: The miRNA was sequenced using llumina Hiseq2500 / Miseq second-generation high-throughput sequencing technology, and the data processing was completed through the processes of removing adapters, removing low-quality, and decontaminating to obtain the final data. The miRNA sequencing raw data was background-corrected by transcriptome data analysis software, and then the t-test was performed to obtain the P value, and then the Fisher test was used to combine the P values to screen for differentially expressed miRNAs.
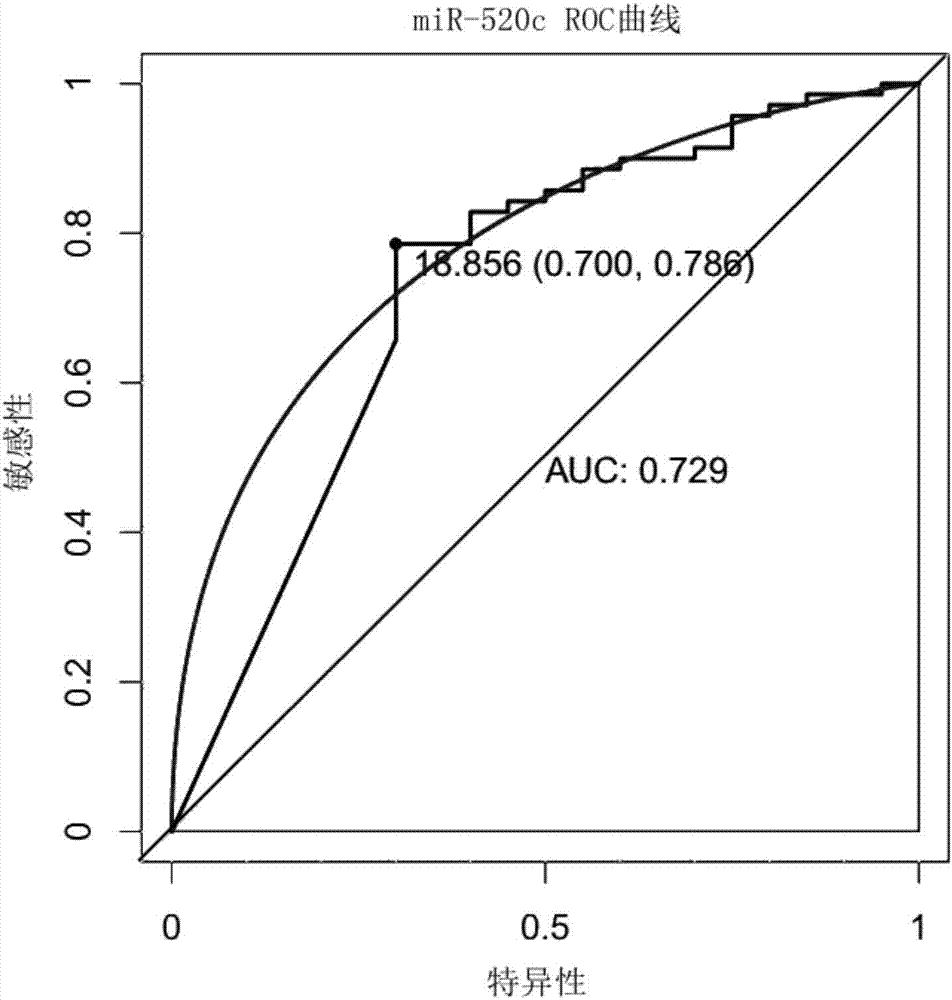
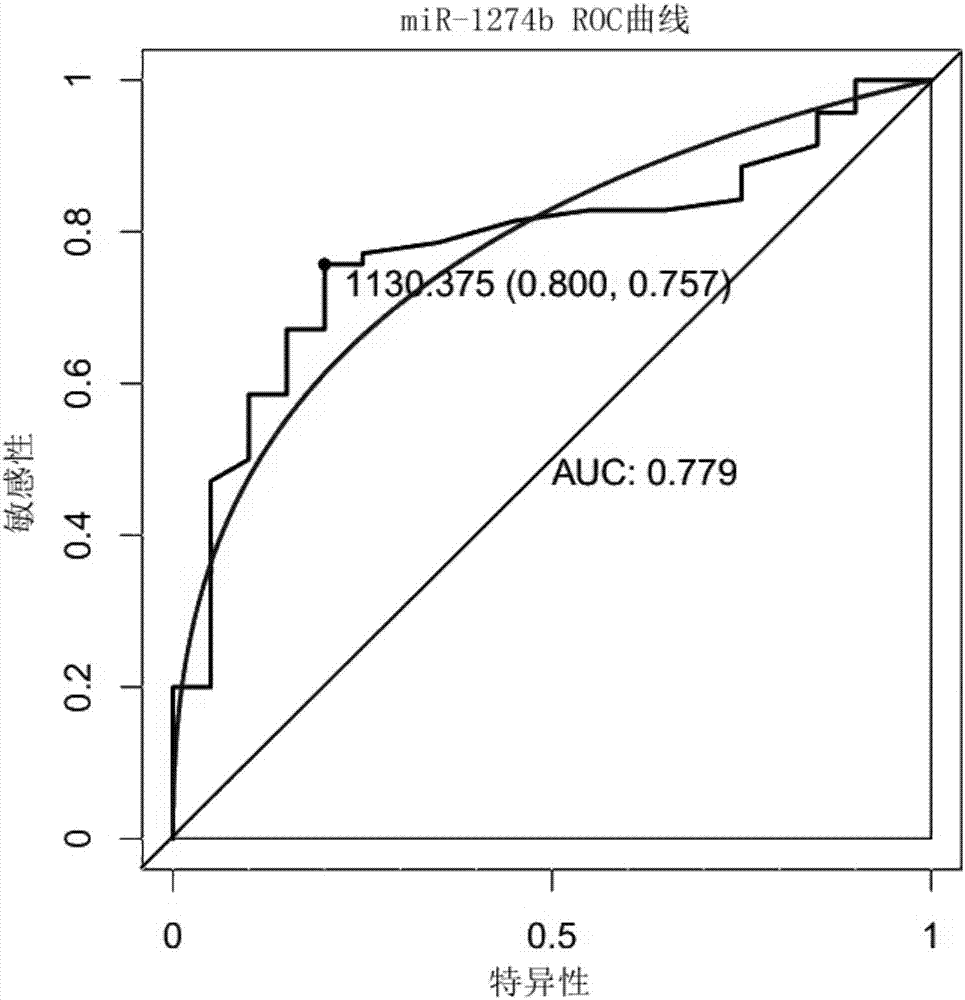
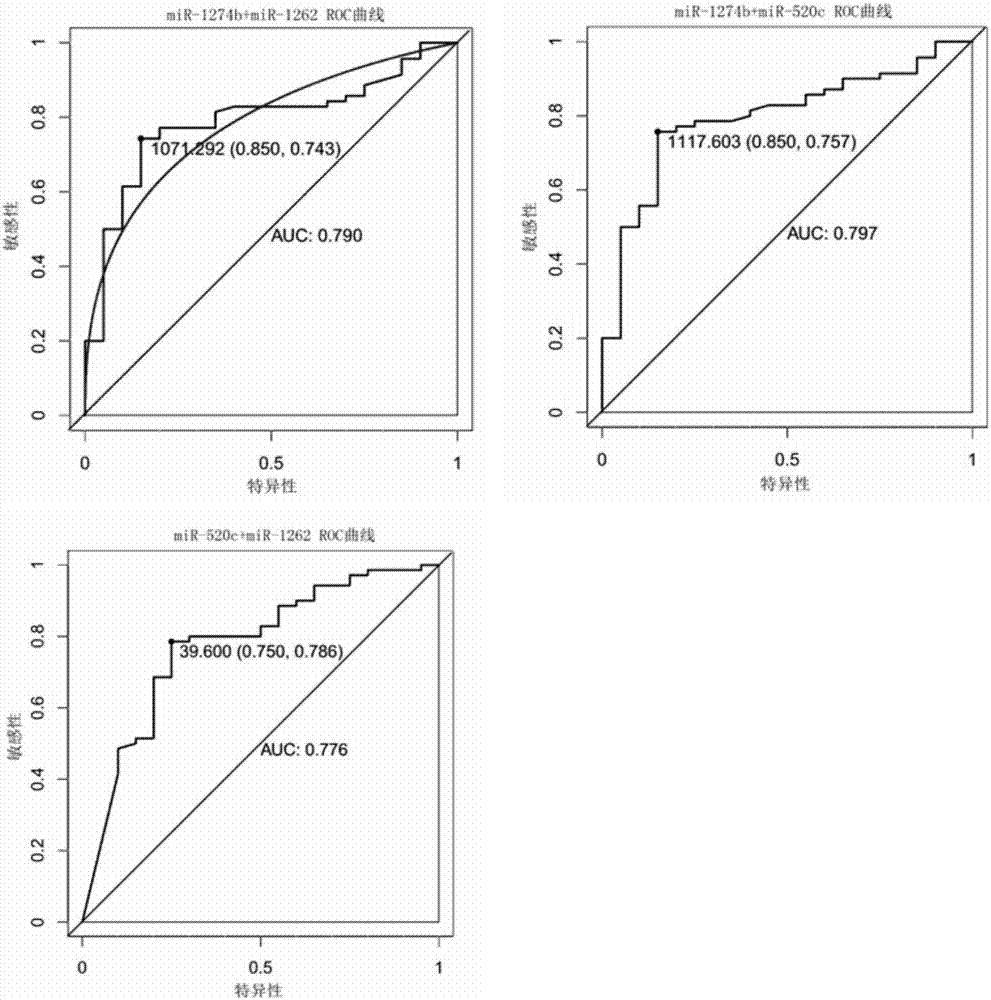
[0056] Finally, miR-1274b, miR-520c, and miR-1262 were selected from the candidate differentially expressed miRNAs for later experimental verification.
Embodiment 3
[0057] Embodiment 3 Electronic Verification
[0058] Electronic verification: 3 sets of mRNA data sets (GSE48060, GSE34198 and GSE61145-GPL6884) and 2 sets of miRNA data sets (GSE61741, GSE31568) were screened from the GEO (Gene Expression Omnibus) database, and 3 sets of mRNA data sets (GSE48060, GSE34198 and GSE61145 -GPL6884) has a total of 13,680 genes. We calculated and merged the effect value by using the metaMA package and using the combined P-value method to obtain 612 (FDR1) were found. The results showed that the electronic verification results were consistent with the expression trend of the sequencing results.
[0059] The method for evaluating the efficacy of a single miRNA molecule or a diagnostic model is to establish a receiver operating characteristic (ROC) curve, and judge the ability of diagnosis by calculating the area under the curve (Area UnderCurve). The value of the area under the ROC curve is between 1.0 and 0.5. In the case of AUC>0.5, the closer the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com