Corn transcription factor ZmbHLH167 and application thereof
A transcription factor, corn technology, applied in the fields of application, genetic engineering, angiosperms/flowering plants, etc., can solve the problem of unobserved mutant phenotypes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
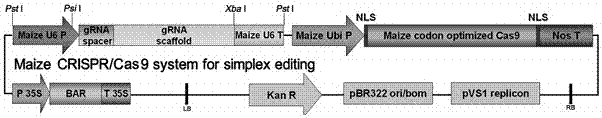
[0029] Example 1: The CRISPR-Cas9 transgenic vector of ZmbHLH167 was constructed and used for transgenic transformation.
[0030] The CRISPR-Cas9 vector suitable for maize that has been constructed in our laboratory was selected as the vector for Agrobacterium transformation of maize immature embryos. Digest the vector with Pst I, and use the synthetic ZmbHLH167 The specific sequence connection guide RNA sequence is inserted into the vector together with the corn U6 promoter and terminator ( figure 1 ), after the sequencing is correct, transform into EHA105 Agrobacterium strain by electric shock. Select the immature embryos of the PBPA corn line 8-12 days after pollination, about 1.5mm in size, as the recipient material, and carry out the transformation of immature embryos. The specific process:
[0031] 1. Agrobacterium infection for 10 minutes-co-cultivation at 20°C for 3 days.
[0032] 2. Recovery culture at 28°C for 7 days-screening culture (bialaphos 1.5 mg / l) at 28°...
Embodiment 2
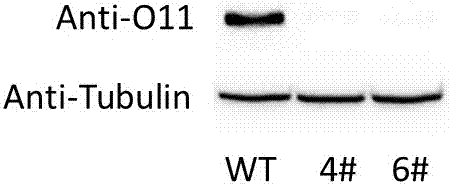
[0038] Embodiment two: ZmbHLH167 Detection of the expression of ZmbHLH167 in the transgenic No. 4 and No. 6 event frameshift mutation materials
[0039] 1. Take 8-10 immature seeds.
[0040] 2. Extract the total protein of ZmbHLH167 mutant material and wild-type material with frameshift mutation respectively. Use liquid nitrogen to grind to powder grade, put into EP tube, add IP lysate, and lyse on ice for 20min.
[0041] 3. Centrifuge at the highest speed and remove the supernatant. Take 4 μl of protein from each of the two samples, add 1 μl of 5×SDS protein loading buffer mixed with 1M DTT, denature at 99°C for 10 minutes, and immediately insert the protein sample on ice.
[0042] 4. SDS-PAGE electrophoresis, the stacking gel is 5%, and after half an hour of 80V electrophoresis, the separating gel is 12.5%, and the electrophoresis time is about 2 hours.
[0043] 5. 200mA transfer film for 1h. Block with 5% milk in TBST for 1 h at room temperature.
[0044] 6. Dilute i...
Embodiment 3
[0050] Embodiment three: ZmbHLH167 Phenotype Observation of CRISPR-Cas9 Event 4 Mutant Kernel
[0051] Observed under natural light and on a light box, respectively ZmbHLH167 The phenotype of the grain of the transgenic frameshift mutant material ( Figure 4 ). Observation ZmbHLH167 Compared with the wild-type grains on the same ear, the grains of transgenic frameshift mutant materials were significantly smaller. Observed on the light box ZmbHLH167 Compared with the wild-type grains, the grains of transgenic frameshift mutant materials showed a significantly lower light transmittance, showing a powdery endosperm phenotype.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com