Avena L. DNA (deoxyribonucleic acid) molecular fingerprint spectrum construction method and application
A technology of DNA molecules and fingerprints, which is applied in the field of molecular biology and plant variety identification, can solve the problem of high development costs and achieve the effect of preventing counterfeit and inferior varieties from entering the market
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 110
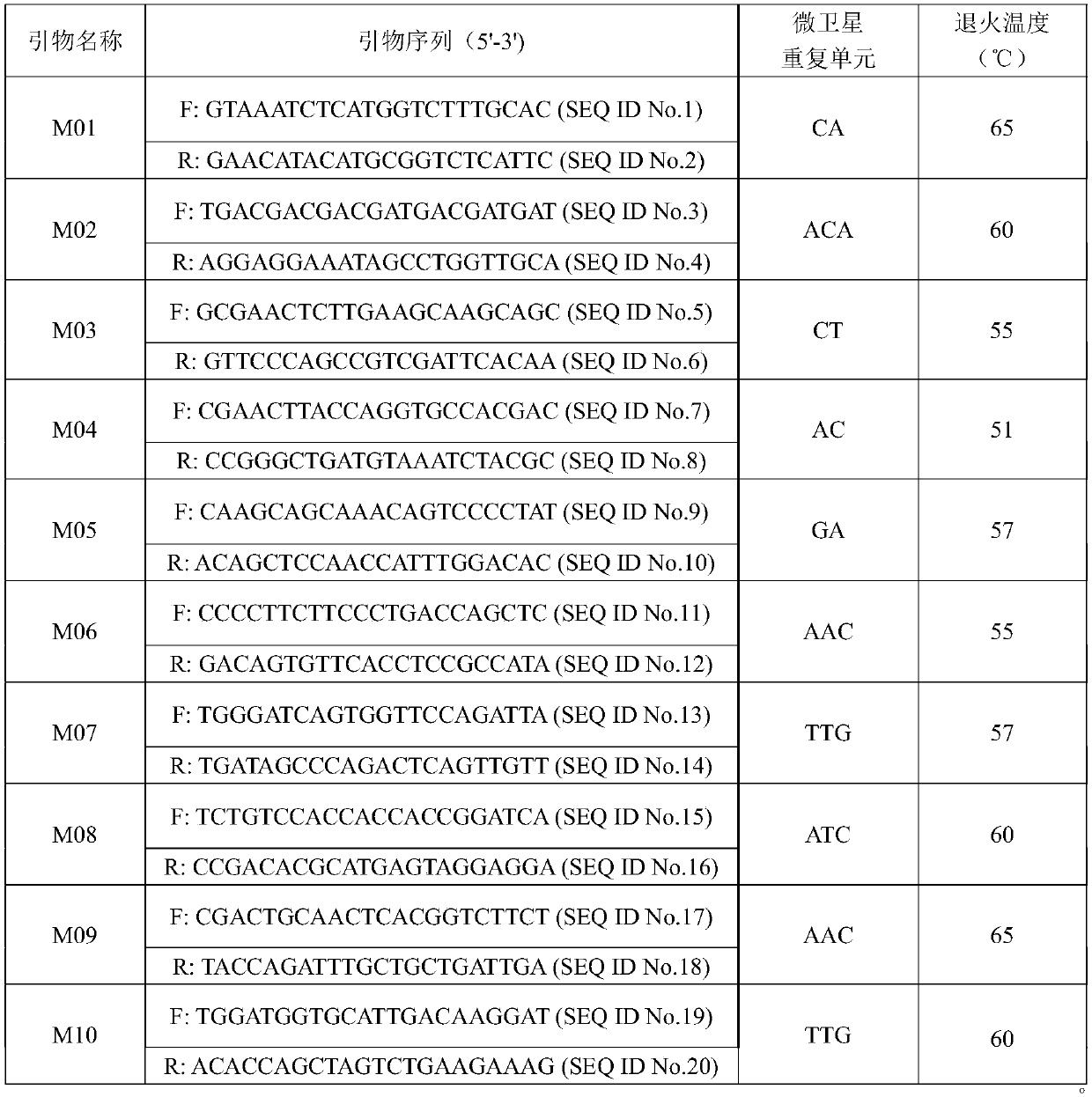
[0030] Example 1 Obtaining 10 pairs of core SSR primers
[0031] In this example, using the genomic DNA of 30 representative oat varieties (from the National Crop Germplasm Resources Preservation Center) in different production areas in my country as templates, by analyzing the number of SSR marker allelic sites, polymorphic information content values and possible Repeatability and other indicators, 10 pairs of SSR primers were screened out, and the molecular marker combinations composed of these primers could completely identify the differences of 30 oat varieties.
[0032] The 30 representative oat varieties are: Pinyan No. 1, Ningyou No. 1, Longyan No. 2, Dingyou No. 1, Bayou No. 1, Bayou No. 9, Baiyan No. 2, Baiyan No. 7, Baiyan No. Yinyan No.1, Caoyou No.1, Huawan No.6, Huazao No.2, Zhangyan No.7, Keyan No.1, Pin No.5, Bayan No.4, Wuzhai Naked Oats, Lijiang Oats, Wuyan No.5 , North China No. 1, Zaoyan No. 1, Jiza No. 2, Hefeng No. 1, Jinyan No. 7, Yanhong No. 10, Tongxi...
Embodiment 2
[0037] Example 2 Construction of Oat Variety Characteristic Fingerprint and Establishment of Oat Variety Identification Method
[0038] 1. Genomic DNA extraction of oat varieties
[0039] The oats were planted in the soil and cultured at room temperature. After two weeks of growth, 0.2 g of the young leaves of each variety of oats were taken in a 1.5 mL centrifuge tube, and liquid nitrogen was added to grind them into powder. The DNA was extracted by the CTAB method, and the experimental steps were carried out according to the instructions of the kit. The concentration of the total DNA was detected by agarose gel electrophoresis and a spectrophotometer, and then the total DNA was diluted to 100 ng / μL and stored at -20°C for future use.
[0040] 2. Using the 30 oat varieties described in Example 1 as materials, use the 10 pairs of SSR primers obtained by screening to carry out PCR amplification of the DNA of each oat variety respectively. The PCR reaction system used in the PC...
Embodiment 3
[0051] Embodiment 3 Application of the method for identifying oat varieties of the present invention
[0052] Take 2 oat varieties (different from the origin in Example 1) as verification materials, use the screened core primers to amplify the 2 oats, and use the method in Example 2 to construct fingerprints for unknown samples. The obtained fingerprints are shown in the table 3.
[0053] Table 3 Fingerprints of varieties to be tested (variety 1 and variety 2)
[0054] name
[0055] Comparing the obtained fingerprint table 3 with the characteristic fingerprint table 2 of the above-mentioned known oat varieties, it is found that the fingerprint codes of the varieties to be tested are the same as those of "Bayou No. 1" and "Baiyan No. 2" in Table 2, indicating that are of the same species as each other.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com