A barley calmodulin gene hvcam1 and its application in salt tolerance
A technology of calmodulin and barley, applied in the application field of barley calmodulin gene HvCAM1 and its salt tolerance, to improve stress resistance, enhance salt tolerance, and improve salt tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
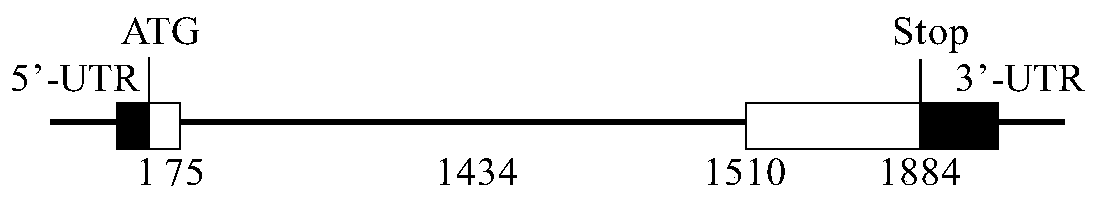
[0030] Embodiment 1 barley calmodulin HvCaM1 gene and promoter cloning
[0031] 1. Processing of plant material
[0032]Barley Golden Promise Seeds with 2% H 2 o 2 After sterilizing for 20 minutes, use ddH 2 After 5 times of washing, the seeds were germinated in the germination box with the ventral side up. Cultivate in the dark in the culture room (14h / 10h, 22°C / 18°C) for 3 days, until the aboveground part is 2cm white, then fill in the light. Seven-day-old barley seedlings were taken, frozen in liquid nitrogen and stored at -70°C for DNA and total RNA extraction.
[0033] 2. Extraction of barley genomic DNA
[0034] Barley genomic DNA was extracted by CTAB method.
[0035] 3. Extraction of barley total RNA
[0036] Total RNA was extracted by Trizol method. Genomic DNA was removed with DNase I enzyme (Takara, Japan).
[0037] 4. Synthesis of cDNA
[0038] Using Takara's The RT reagent Kit Perfect Real Time (DRR037A) kit was used to reverse transcribe the total RNA...
Embodiment 2
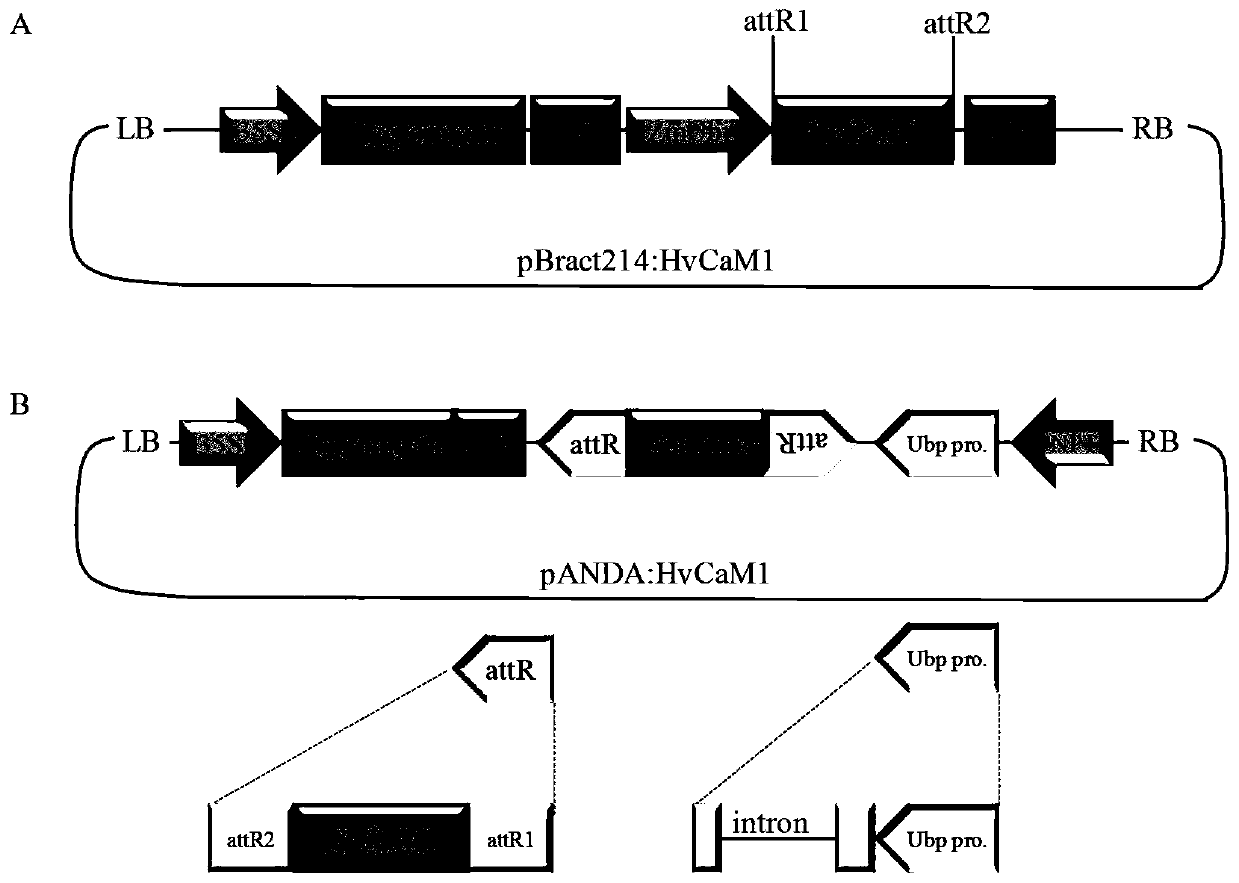
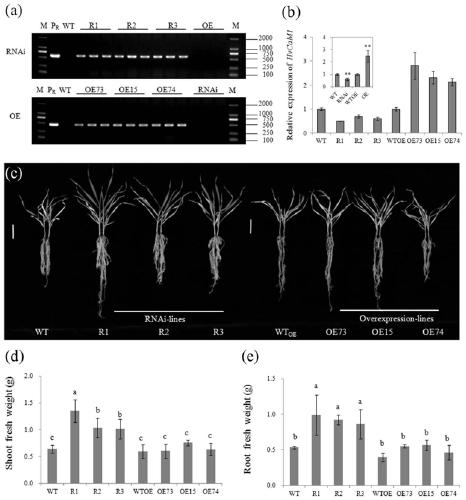
[0058] Example 2 RNAi and overexpression transgenic methods verify HvCaM1 gene function
[0059] 1. Construction of pBract214: HvCaM1 overexpression vector
[0060] Adopt the method of Gateway (Invitrogen, the U.S.), at first take the positive T vector containing cDNA in the above-mentioned embodiment 1 as template, design OE-HvCaM1-F / R primer amplification length is to cover the 450bp fragment of HvCaM1 coding region, primer information for:
[0061] OE-HvCaM1-F:5'-ATGGCGGACCAGCTCACC-3', (SEQ ID No: 11)
[0062] OE-HvCaM1-R:5'-TCACTTGGCCATCATGACC-3'. (SEQ ID No: 12)
[0063] After two rounds of PCR amplification, the BP Clonase TM II Enzyme Mix kit (Cat.No.11789-020, Invitrogen, U.S.) imports the fragment into pDONR TM In the / zeo vector (LifeTechnology), Escherichia coli DH5α was transformed and sent to the company for sequencing. Select the positive carrier containing the target fragment with correct sequencing, and then pass LR Clonase TM II Enzyme Mix Kit (Ca...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com