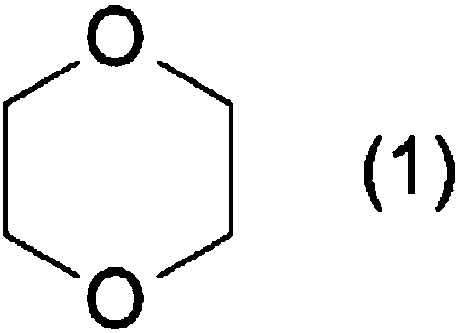
Primer set for detecting 1,4-dioxane degrading bacterium, and method for detecting and quantifying 1,4-dioxane degrading bacterium
A detection method and primer set technology, applied in the fields of botanical equipment and methods, biochemical equipment and methods, applications, etc., can solve the problems of time-consuming and uncertain cultivation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] "Example 1" Identification of the genome involved in 1,4-dioxane degradation
[0036] Predict the open reading frame (ORF) from the draft genome data of the D17 strain and select the annotated ORF through BLAST search (BLASTP), and use the eArray system of Agilent Technology to design the DNA probe, thereby designing 7511 A sequence of probes. The 8×15K format of Agilent Technology was used to make a custom DN Amicroarray slide.
[0037] In an inorganic salt medium supplemented with 500 mg / L of 1,4-dioxane (1g / L K 2 HPO 4 , 1g / L(N H 4 ) 2 SO 4 , 50mg / L NaCl, 200mg / L MgSO 4 ·7H 2 O, 10mg / L FeCl 3 , 50mg / L Ca Cl 2 , PH 7.0) was inoculated with colonies of strain D17, and cultured with rotation and shaking at 28°C and 120 rpm. During the culture period, the concentration of 1,4-dioxane in the culture solution was measured over time. As a result, the concentration of 1,4-dioxane decreased by about 40% after 6 days and decreased by about 60% after 7 days. Therefore, a part of t...
Embodiment 2
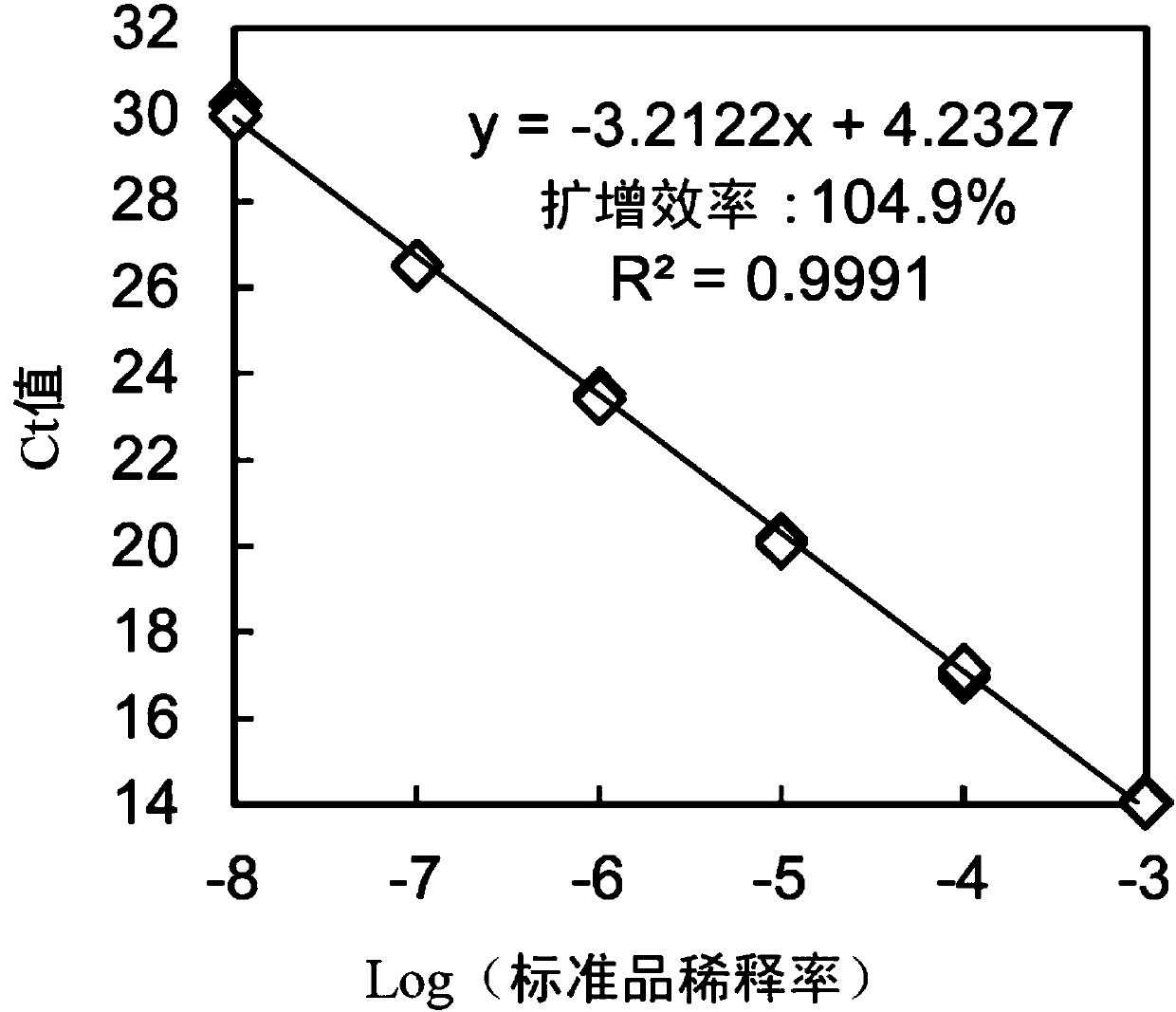
[0043] "Example 2" Quantification of bacterial cell mass by real-time PCR method targeting thm genome
[0044] The base sequence of the thm genome was identified from the draft genome data of the D17 strain, using the Primer3 program (T. KORESSAAR, M. REMM: Bioinformatics, 23, 10, 1289-1291 (2007) "Enhanc ements and modifications of primer design program Primer3", A.UNTERGASS ER, I.CUTCUTACHE, T.KORESSAAR, J.YE, BCFAIRCLOTH, M.REMM, SGROZEN: Nucleic Acids Res. 40, 15, e115 (2012) "Primer3-new capabilities and interfaces") selected Candidate primers for real-time PCR. Next, the Primer-Blast program (J.YE, G.COULOURIS, I.ZARETSKAYA, I.CUTCUTACHE, S.ROZEN, TLMADDEN: BMC Bioinformatics, 13, 134 (2012) "Primer-BLAST: Atool to design target-specif ic primers for polymerase chain reaction”) Search for the homology between the sequence of the candidate primer and the GenBank registered sequence to select the primer with high specificity. The selected primers with high specificity were ...
Embodiment 3
[0056] "Example 3"
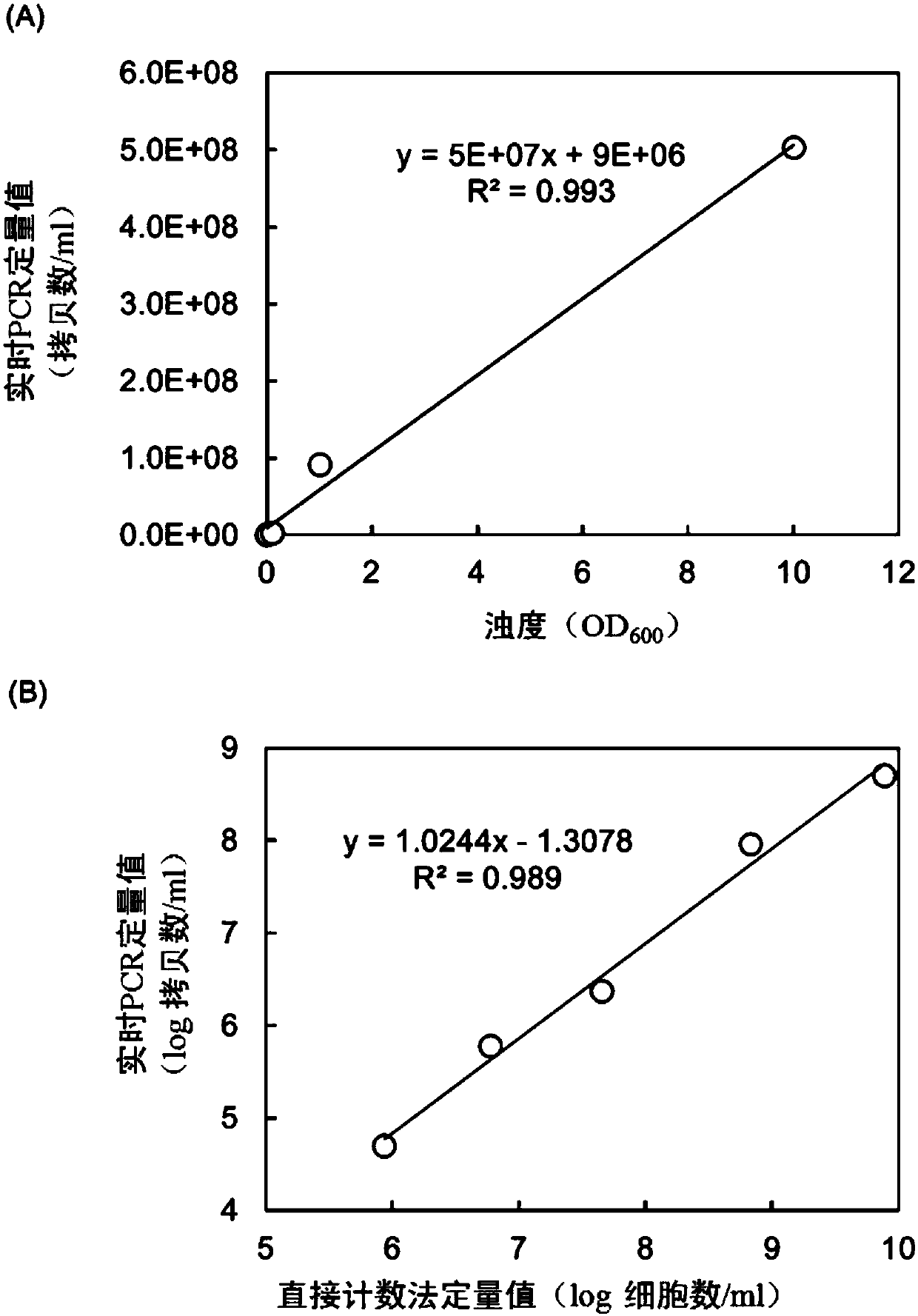
[0057] For the D17 strain, a primer containing the base sequence shown in SEQ ID NO: 1 (5'-TGATTATGTGGG GCTGGTTATG-3') and a reverse primer containing the base sequence shown in SEQ ID NO: 2 (5'-CGAGGAAAGTTGTGTTCGTGATG) -3') The quantitative value and turbidity (OD 600 The relationship between the measured value) and the quantitative value of the direct counting method (A ODC) using acridine orange was investigated.
[0058] The D17 strain was cultured in MGY medium (malt extract: 10 g / L, glucose: 4 g / L, yeast extract: 4 g / L) for 2 weeks. Afterwards, the bacteria were recovered by centrifugal separation, washed twice with physiological saline and made into different turbidity (OD 600 ) Cell solution, which can be used for AODC and real-time PCR. In addition, the OD of each cell solution 600 The values are 0.00015, 0.0089, 0.1058, 1.01305 and 10.01145.
[0059]
[0060] 2.29 mL of stain preservation solution (NaCl: 1.75%, acridine orange: 0.005%, glutaral dehy...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap