Specific human gene fragment, primer and probe for detecting specific human gene fragment and application of specific human gene fragment
A gene fragment and probe technology, used in the field of pharmacokinetic biodetection, can solve the problems of inability to correspond to cell mass, high non-specific ligation products, difficult absolute quantification of qPCR, etc., and achieve accurate and sensitive detection, strong sensitivity, Accurate effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 Acquisition of Specific Human Gene Fragments
[0036] 1.1 Acquisition of the full-length sequence of the target gene
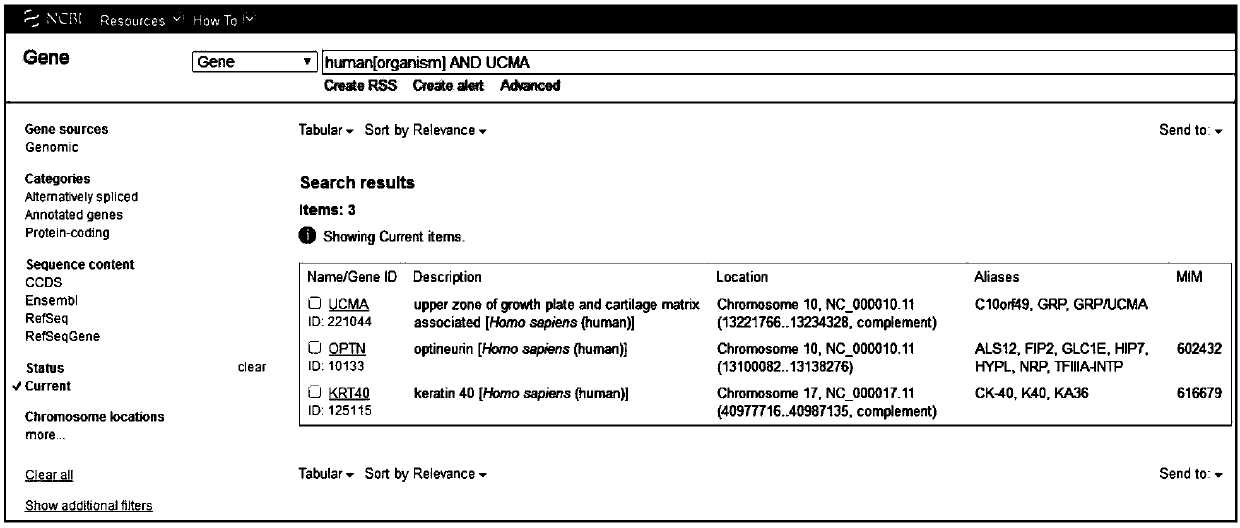
[0037] In Genome, a sub-site of the NCBI website, search for possible specific gene names of human origin, such as figure 1 As shown, click on GenBank to obtain the full-length sequence of the target gene on the genome of different species.
[0038] 1.2 Comparison of target genes among different species
[0039] Also on the NCBI website, use the nucleic acid comparison function in its sub-site BLAST to input the genome sequences of the same target gene in different species, such as figure 2 As shown, after the comparison analysis, the sequence difference information of the target gene among different species can be obtained.
[0040] 1.3 Selection of specific human gene fragments
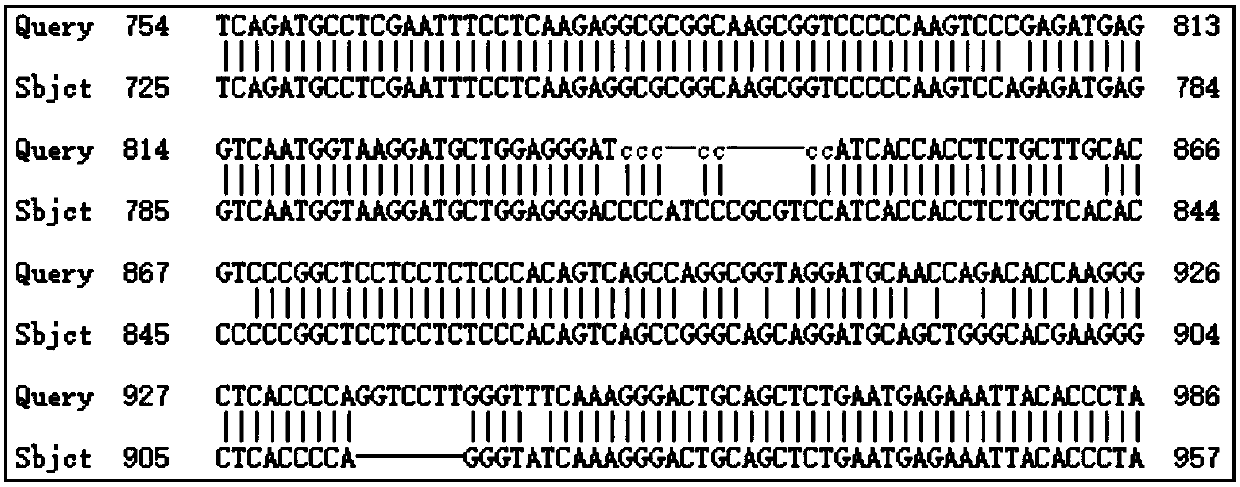
[0041]On the basis of the comparison analysis in 1.2, find the region with the greatest difference between the human gene and other animal gene sequences, such...
Embodiment 2
[0042] Design synthesis and verification of embodiment 2 primer probe
[0043] 2.1 Design of human-specific primer probes
[0044] Use the software Premier primer 5 to design high-quality primer probes on the selected target fragments. The general principle is that the primer sequence is 18-25bp, the Tm value is 50-60°C, and the GC content is 40-60%; the corresponding probe is at the end close to the primer, and the Tm value is about 10°C higher than that of the primer.
[0045] 2.2 Theoretical analysis of human-specific primers and probes
[0046] First, input the designed pair of primers into the Primer BLAST under the BLAST sub-site of the NCBI website, such as Figure 4 As shown, then, select the properties of the primers to be detected, such as on the genome, which species, etc., such as Figure 5 As shown; click Getprimer to obtain the detection results. It can be seen that in many animal species, the designed primers can only specifically recognize the human genome s...
Embodiment 3
[0049] The construction of embodiment 3 standard plasmid and the drafting of standard curve
[0050] 3.1 Construct a standard plasmid containing the target sequence:
[0051] The theoretical analysis and experimental test results in Example 2 prove that the target sequence can be used to specifically detect human cells (genome fragments), therefore, the target sequence can be connected to the target vector to construct a standard plasmid.
[0052] 3.1.1 Synthetic primers:
[0053] Sense: GTAGGTCGTATATTGGATTG, namely the base sequence of primer-F shown in SEQ ID NO.2.
[0054] Anti-sense: AGGTGTGTTTAATGTAGC, namely the base sequence of primer-R shown in SEQ ID NO.3.
[0055] 3.1.2 Clone target sequence (BIO-RAD PCR instrument):
[0056] Reaction system: PrimeStar MAX(25ul) / 5umol Primers(F+R)2ul / human genome DNA(200ng) / H2O up to 50ul
[0057] Reaction conditions: 98°C for 5min, 98°C for 15s, 49°C for 30s, 72°C for 20s, 12°C forever, 36 cycles
[0058] 3.1.3 Running glue cut...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com