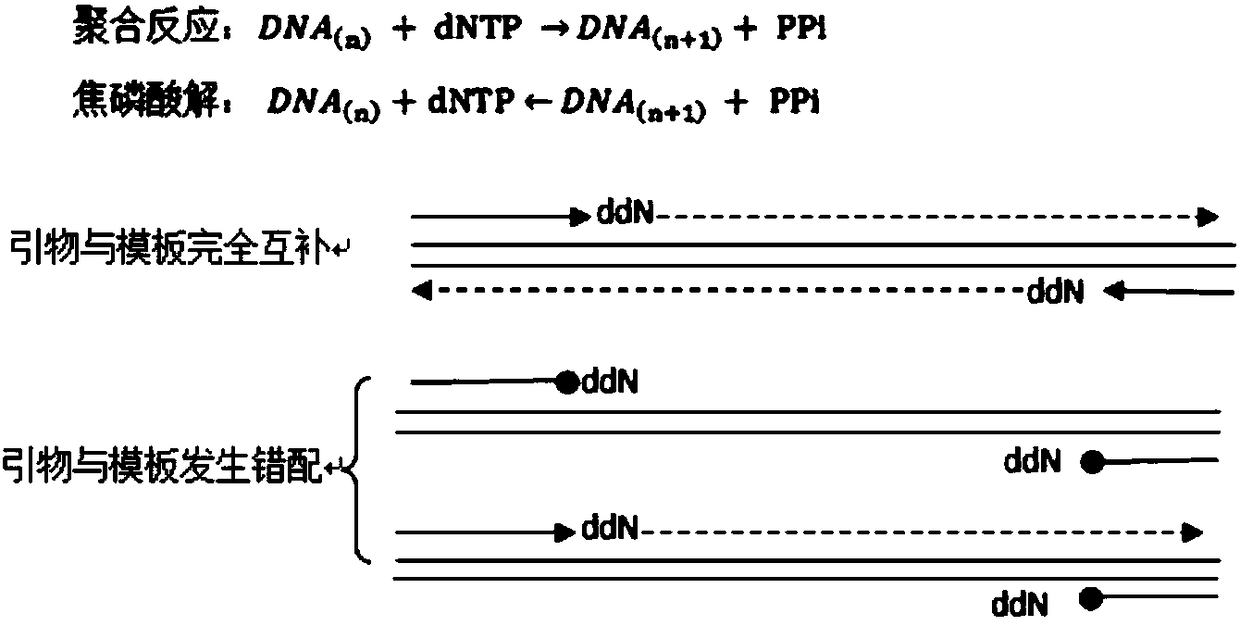
High-specificity multiplex-PCR (polymerase chain reaction) detection method for base mutation
A technology with high specificity and detection method, which is applied in the field of multiplex PCR detection of high specificity base mutation, to achieve the effect of improving detection specificity and sensitivity, high efficiency, and improving tissue heterogeneity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] To verify the gene mutation shown by the NGS data of a cancer patient, design 5 pairs of primers (see Table 4), and mix all the primers in equal proportions to form a primer combination.
[0071] The primer information table in the embodiment 1 of table 4
[0072]
[0073]
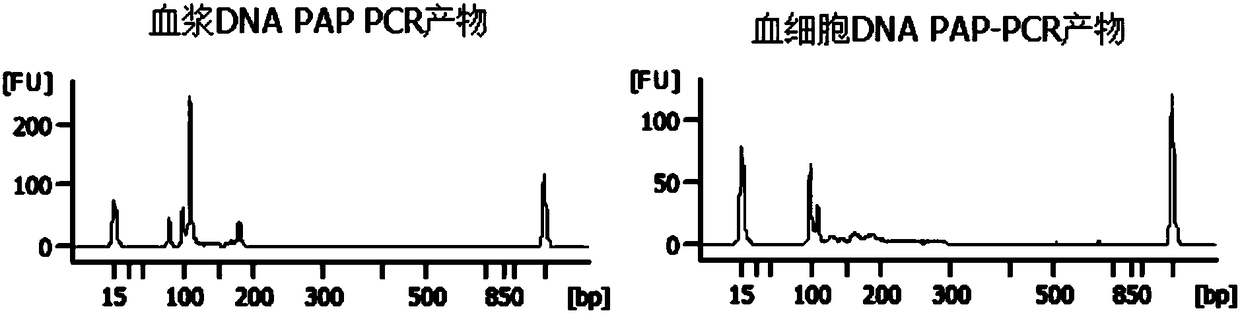
[0074] Use 5 pairs of primer combinations to carry out multiplex PCR on the patient's plasma DNA and blood cell DNA respectively (for the detection results of PCR products, see figure 2 ), after which a high-throughput sequencing library was constructed, and the sequencing results showed that the patient's JAK2 and DNMT3A genes were mutated, and the gene frequencies were 0.9% and 0.7% respectively (see Table 5).
[0075] Table 5 The sequencing results of Example 1
[0076] Chr.
Embodiment 2
[0078] To verify the gene mutation shown by the NGS data of a cancer patient, 4 pairs of primer pairs were designed (see Table 6), and all primers were mixed in equal proportions to form a primer combination.
[0079] The primer information table in the table 6 embodiment 2
[0080]
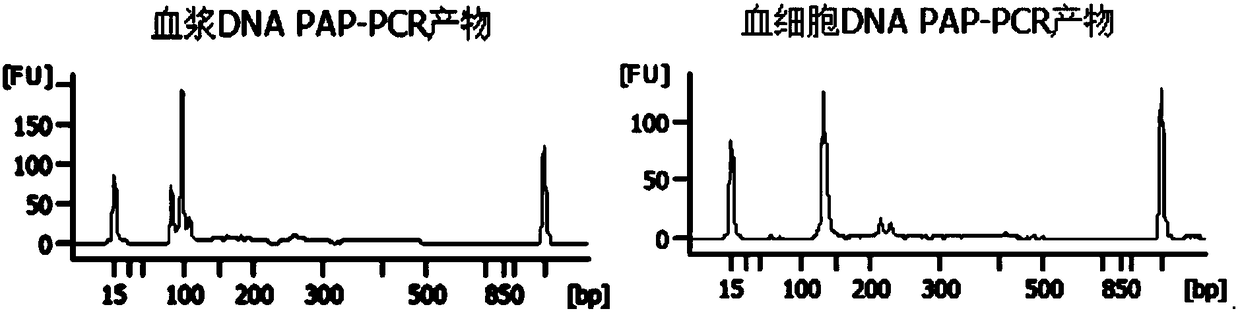
[0081] Use 4 pairs of primer combinations to carry out multiplex PCR on plasma DNA and blood cell DNA respectively (for the detection results of PCR products, see image 3 ), then a high-throughput sequencing library was constructed, and the sequencing results showed that the patient's DNMT3A gene was mutated, with a gene frequency of 0.85% (see Table 7).
[0082] Table 7 The sequencing results of Example 2
[0083]
Embodiment 3
[0085] 6 pairs of primers (see Table 9) were designed for hot spot mutations (see Table 8) of lung cancer with drug guidance significance, and all primers were mixed in equal proportions to form a primer combination. Multiplex PCR was performed on preoperative plasma DNA, blood cell DNA, and cancer tissue DNA from a patient with stage II non-small cell lung cancer. The product detection results are shown in Figure 4 , Hiseq sequencing results showed that relative to blood cell DNA, EGFR 19del somatic mutations appeared in both cancer tissue and plasma of the patient, and the gene frequency detection values were 1.2% and 0.8%, respectively.
[0086] Table 8 Hotspot mutations in lung cancer
[0087]
[0088]
[0089]
[0090] Primer information table in table 9 embodiment 3
[0091]
[0092]
[0093] As an improvement or replacement of the present invention, the 5' end of the modified primer can be designed to connect the complementary universal sequence of th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com