A high-throughput method for screening and detecting transgenic elements
A detection method and genetic element technology, applied in the field of molecular biology, to achieve the effects of reducing the probability of error, stabilizing the detection method, and saving costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1 Application of the method of the present invention detects whether the corn sample contains NOS Terminator Transgenic Components
[0047] 1. Preparation of DNA template
[0048] Using the DNA extraction method of TPS or CTAB, DNA was extracted from 101 corn leaves as templates for PCR reactions. Prepare the template for the control at the same time: the positive control contains NOS Terminator transgenic maize and containing NOS The plasmid of the terminator, the negative control is non-transgenic corn, and the blank control is sterile ultrapure water.
[0049] 2. Design and synthesis of primers
[0050] Designed according to the sequence of gene fragments in Genbank NOS terminators (Table 1) and maize endogenous genes ZSSIIb Specific amplification primers (Table 2). exist NOS Adding a FAM tag sequence to the 5' end of the terminator forward primer becomes "GAAGGTGACCAAGTTCATGCTCAGTGGTCCCAAAGATGGAC"; in ZSSIIb A HEX tag sequence was added to the ...
Embodiment 2
[0062] The pairing combination of embodiment 2 primers
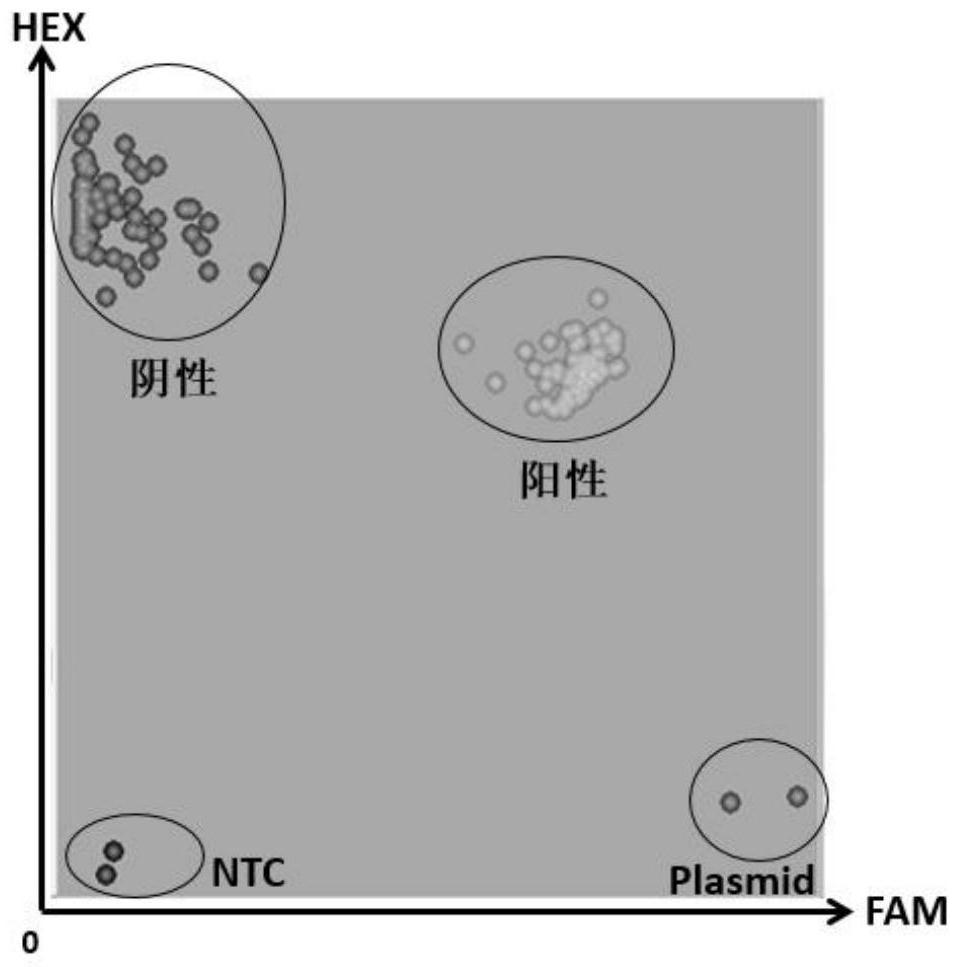
[0063] When using the method of the present invention for detection, a pair of transgenic element primers and a pair of endogenous gene primers are added to the reaction system. How to ensure that the primers of the two genes do not interfere with each other becomes a difficulty in this application. Through experiments, a series of primer pairs were screened out (Table 5). These paired primers were validated against known negative and positive samples. The specific experimental steps are as follows:
[0064] (1) Prepare DNA templates: prepare DNA from non-transgenic samples, plant sample DNA containing transgenic elements, and E. coli plasmid DNA containing transgenic elements.
[0065] (2) Construct the reaction system: transgenic element primer (FAM tag sequence added to the 5' end of the forward primer), endogenous gene primer (HEX tag sequence added to the 5' end of the forward primer), KASP Master Mix, and DNA tem...
Embodiment 3
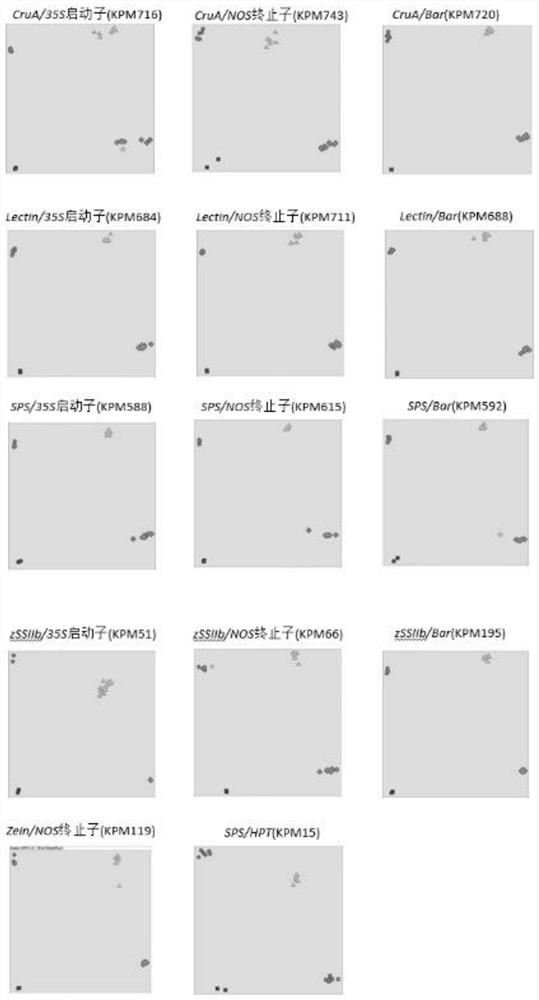
[0070] Example 3 The accuracy of high-throughput detection using the method of the present invention.
[0071] When using the method of the present invention to detect a large number of samples, it becomes a difficult point in this application to clearly distinguish positive and negative samples on the scatter diagram of the scanning results. In order to determine the effect of using the method of the present invention to carry out high-throughput detection on a large number of samples, 384 known negative and positive samples are detected with KPM743, KPM119, and KPM588 in Table 5 as examples, and the detection results are compared with the genotypes of the samples themselves , to verify the accuracy of the method for detecting a large number of samples. The specific experimental steps are as follows:
[0072] (1) Prepare DNA template: prepare DNA from non-transgenic plant samples, containing 35S promoter or NOS DNA from plant samples for terminators, E. coli plasmids conta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com